The human gut bacterial genotoxin colibactin alkylates DNA
- PMID: 30765538
- PMCID: PMC6407708
- DOI: 10.1126/science.aar7785
The human gut bacterial genotoxin colibactin alkylates DNA
Abstract
Certain Escherichia coli strains residing in the human gut produce colibactin, a small-molecule genotoxin implicated in colorectal cancer pathogenesis. However, colibactin's chemical structure and the molecular mechanism underlying its genotoxic effects have remained unknown for more than a decade. Here we combine an untargeted DNA adductomics approach with chemical synthesis to identify and characterize a covalent DNA modification from human cell lines treated with colibactin-producing E. coli Our data establish that colibactin alkylates DNA with an unusual electrophilic cyclopropane. We show that this metabolite is formed in mice colonized by colibactin-producing E. coli and is likely derived from an initially formed, unstable colibactin-DNA adduct. Our findings reveal a potential biomarker for colibactin exposure and provide mechanistic insights into how a gut microbe may contribute to colorectal carcinogenesis.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
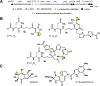

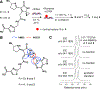

Comment in
-
Revealing a microbial carcinogen.Science. 2019 Feb 15;363(6428):689-690. doi: 10.1126/science.aaw5475. Science. 2019. PMID: 30765550 No abstract available.
References
-
- Lynch SV, Pedersen O, The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016). - PubMed
-
- Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E, Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006). - PubMed
-
- Arthur JC, Peréz-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C, Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

