How a circularized tmRNA moves through the ribosome
- PMID: 30765567
- PMCID: PMC6440651
- DOI: 10.1126/science.aav9370
How a circularized tmRNA moves through the ribosome
Abstract
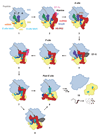
During trans-translation, transfer-messenger RNA (tmRNA) and small protein B (SmpB) together rescue ribosomes stalled on a truncated mRNA and tag the nascent polypeptide for degradation. We used cryo-electron microscopy to determine the structures of three key states of the tmRNA-SmpB-ribosome complex during trans translation at resolutions of 3.7 to 4.4 angstroms. The results show how tmRNA and SmpB act specifically on stalled ribosomes and how the circularized complex moves through the ribosome, enabling translation to switch from the old defective message to the reading frame on tmRNA.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

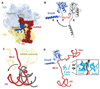

References
-
- Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13:285–297. - PubMed
-
- Moore SD, Sauer RT. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol Microbiol. 2005;58:456–466. - PubMed
-
- Chadani Y, et al. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol. 2010;78:796–808. - PubMed
-
- Chadani Y, Ono K, Kutsukake K, Abo T. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol Microbiol. 2011;80:772–785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

