Numerous cultivated and uncultivated viruses encode ribosomal proteins
- PMID: 30765709
- PMCID: PMC6375957
- DOI: 10.1038/s41467-019-08672-6
Numerous cultivated and uncultivated viruses encode ribosomal proteins
Abstract
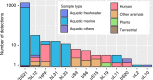
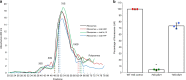
Viruses modulate ecosystems by directly altering host metabolisms through auxiliary metabolic genes. However, viral genomes are not known to encode the core components of translation machinery, such as ribosomal proteins (RPs). Here, using reference genomes and global-scale viral metagenomic datasets, we identify 14 different RPs across viral genomes arising from cultivated viral isolates and metagenome-assembled viruses. Viruses tend to encode dynamic RPs, easily exchangeable between ribosomes, suggesting these proteins can replace cellular versions in host ribosomes. Functional assays confirm that the two most common virus-encoded RPs, bS21 and bL12, are incorporated into 70S ribosomes when expressed in Escherichia coli. Ecological distribution of virus-encoded RPs suggests some level of ecosystem adaptations as aquatic viruses and viruses of animal-associated bacteria are enriched for different subsets of RPs. Finally, RP genes are under purifying selection and thus likely retained an important function after being horizontally transferred into virus genomes.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

