Efficient neural decoding of self-location with a deep recurrent network
- PMID: 30768590
- PMCID: PMC6407788
- DOI: 10.1371/journal.pcbi.1006822
Efficient neural decoding of self-location with a deep recurrent network
Abstract
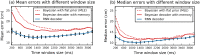
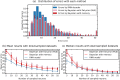
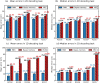
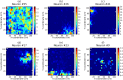
Place cells in the mammalian hippocampus signal self-location with sparse spatially stable firing fields. Based on observation of place cell activity it is possible to accurately decode an animal's location. The precision of this decoding sets a lower bound for the amount of information that the hippocampal population conveys about the location of the animal. In this work we use a novel recurrent neural network (RNN) decoder to infer the location of freely moving rats from single unit hippocampal recordings. RNNs are biologically plausible models of neural circuits that learn to incorporate relevant temporal context without the need to make complicated assumptions about the use of prior information to predict the current state. When decoding animal position from spike counts in 1D and 2D-environments, we show that the RNN consistently outperforms a standard Bayesian approach with either flat priors or with memory. In addition, we also conducted a set of sensitivity analysis on the RNN decoder to determine which neurons and sections of firing fields were the most influential. We found that the application of RNNs to neural data allowed flexible integration of temporal context, yielding improved accuracy relative to the more commonly used Bayesian approaches and opens new avenues for exploration of the neural code.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- O’keefe J, Nadel L. The hippocampus as a cognitive map. Oxford: Clarendon Press; 1978.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

