Huntingtin associates with the actin cytoskeleton and α-actinin isoforms to influence stimulus dependent morphology changes
- PMID: 30768638
- PMCID: PMC6377189
- DOI: 10.1371/journal.pone.0212337
Huntingtin associates with the actin cytoskeleton and α-actinin isoforms to influence stimulus dependent morphology changes
Abstract
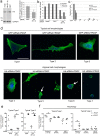
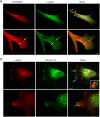
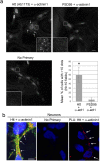
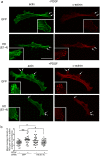
One response of cells to growth factor stimulus involves changes in morphology driven by the actin cytoskeleton and actin associated proteins which regulate functions such as cell adhesion, motility and in neurons, synaptic plasticity. Previous studies suggest that Huntingtin may be involved in regulating morphology however, there has been limited evidence linking endogenous Huntingtin localization or function with cytoplasmic actin in cells. We found that depletion of Huntingtin in human fibroblasts reduced adhesion and altered morphology and these phenotypes were made worse with growth factor stimulation, whereas the presence of the Huntington's Disease mutation inhibited growth factor induced changes in morphology and increased numbers of vinculin-positive focal adhesions. Huntingtin immunoreactivity localized to actin stress fibers, vinculin-positive adhesion contacts and membrane ruffles in fibroblasts. Interactome data from others has shown that Huntingtin can associate with α-actinin isoforms which bind actin filaments. Mapping studies using a cDNA encoding α-actinin-2 showed that it interacts within Huntingtin aa 399-969. Double-label immunofluorescence showed Huntingtin and α-actinin-1 co-localized to stress fibers, membrane ruffles and lamellar protrusions in fibroblasts. Proximity ligation assays confirmed a close molecular interaction between Huntingtin and α-actinin-1 in human fibroblasts and neurons. Huntingtin silencing with siRNA in fibroblasts blocked the recruitment of α-actinin-1 to membrane foci. These studies support the idea that Huntingtin is involved in regulating adhesion and actin dependent functions including those involving α-actinin.
Conflict of interest statement
Kimberly Kegel-Gleason is on the editorial board of PLOS ONE. KKG’s spouse owns less than 0.1% stock in the following companies: Bristol-Myers Squibb Company, Cisco Systems, Inc., GE Healthcare Life Sciences, Generex Biotechnology Corporation, GlaxoSmithKline, Metabolix Bioplastics, Nanogen, Inc., Nanometrics, inc., StemCells, Inc. All purchases are compliant with Research Procurement standards that both individual and organizational conflicts of interest compliance with Uniform Guidance (UG) Procurement Standards issued by the Office of Management and Budget (OMB). This does not alter our adherence to PLOS ONE policies on sharing data and materials. No other authors have conflicts of interest to report. All other authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- HDCRG. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

