A H2AX⁻CARP-1 Interaction Regulates Apoptosis Signaling Following DNA Damage
- PMID: 30769864
- PMCID: PMC6406907
- DOI: 10.3390/cancers11020221
A H2AX⁻CARP-1 Interaction Regulates Apoptosis Signaling Following DNA Damage
Abstract
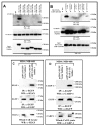
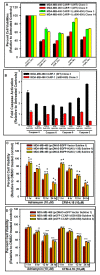
Cell Cycle and Apoptosis Regulatory Protein (CARP-1/CCAR1) is a peri-nuclear phosphoprotein that regulates apoptosis via chemotherapeutic Adriamycin (doxorubicin) and a novel class of CARP-1 functional mimetic (CFM) compounds. Although Adriamycin causes DNA damage, data from Comet assays revealed that CFM-4.16 also induced DNA damage. Phosphorylation of histone 2AX (γH2AX) protein is involved in regulating DNA damage repair and apoptosis signaling. Adriamycin or CFM-4.16 treatments inhibited cell growth and caused elevated CARP-1 and γH2AX in human breast (HBC) and cervical cancer (HeLa) cells. In fact, a robust nuclear or peri-nuclear co-localization of CARP-1 and γH2AX occurred in cells undergoing apoptosis. Knock-down of CARP-1 diminished γH2AX, their co-localization, and apoptosis in CFM-4.16- or Adriamycin-treated cells. We found that CARP-1 directly binds with H2AX, and H2AX interacted with CARP-1, but not CARP-1 (Δ600⁻652) mutant. Moreover, cells expressing CARP-1 (Δ600⁻652) mutant were resistant to apoptosis, and had diminished levels of γH2AX, when compared with cells expressing wild-type CARP-1. Mutagenesis studies revealed that H2AX residues 1⁻35 harbored a CARP-1-binding epitope, while CARP-1 amino acids 636⁻650 contained an H2AX-interacting epitope. Surface plasmon resonance studies revealed that CARP-1 (636⁻650) peptide bound with H2AX (1⁻35) peptide with a dissociation constant (Kd) of 127 nM. Cells expressing enhanced GFP (EGFP)-tagged H2AX (1⁻35) peptide or EGFP-tagged CARP-1 (636⁻650) peptide were resistant to inhibition by Adriamycin or CFM-4.16. Treatment of cells with transactivator of transcription (TAT)-tagged CARP-1 (636⁻650) peptide resulted in a moderate, statistically significant abrogation of Adriamycin-induced growth inhibition of cancer cells. Our studies provide evidence for requirement of CARP-1 interaction with H2AX in apoptosis signaling by Adriamycin and CFM compounds.
Keywords: CCAR1/CARP-1; apoptosis; cancer cells; chemotherapeutics; γH2AX.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Rishi A.K., Zhang L., Boyanapalli M., Wali A., Mohammad R.M., Yu Y., Fontana J.A., Hatfield J.S., Dawson M.I., Majumdar A.P.N., et al. Identification and characterization of a Cell-Cycle and Apoptosis Regulatory Protein (CARP)-1 as a novel mediator of apoptosis signaling by Retinoid CD437. J. Biol. Chem. 2003;278:33422–33435. doi: 10.1074/jbc.M303173200. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

