The search for a PKR code-differential regulation of protein kinase R activity by diverse RNA and protein regulators
- PMID: 30770398
- PMCID: PMC6467004
- DOI: 10.1261/rna.070169.118
The search for a PKR code-differential regulation of protein kinase R activity by diverse RNA and protein regulators
Abstract
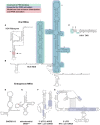
The interferon-inducible protein kinase R (PKR) is a key component of host innate immunity that restricts viral replication and propagation. As one of the four eIF2α kinases that sense diverse stresses and direct the integrated stress response (ISR) crucial for cell survival and proliferation, PKR's versatile roles extend well beyond antiviral defense. Targeted by numerous host and viral regulators made of RNA and proteins, PKR is subject to multiple layers of endogenous control and external manipulation, driving its rapid evolution. These versatile regulators include not only the canonical double-stranded RNA (dsRNA) that activates the kinase activity of PKR, but also highly structured viral, host, and artificial RNAs that exert a full spectrum of effects. In this review, we discuss our deepening understanding of the allosteric mechanism that connects the regulatory and effector domains of PKR, with an emphasis on diverse structured RNA regulators in comparison to their protein counterparts. Through this analysis, we conclude that much of the mechanistic details that underlie this RNA-regulated kinase await structural and functional elucidation, upon which we can then describe a "PKR code," a set of structural and chemical features of RNA that are both descriptive and predictive for their effects on PKR.
Keywords: PKR; allosteric regulation; antiviral defense; innate immunity; noncoding RNA.
Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
