Recessive mutations in muscle-specific isoforms of FXR1 cause congenital multi-minicore myopathy
- PMID: 30770808
- PMCID: PMC6377633
- DOI: 10.1038/s41467-019-08548-9
Recessive mutations in muscle-specific isoforms of FXR1 cause congenital multi-minicore myopathy
Abstract
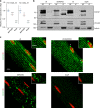
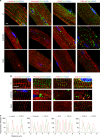
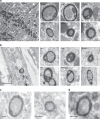
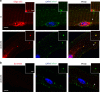
FXR1 is an alternatively spliced gene that encodes RNA binding proteins (FXR1P) involved in muscle development. In contrast to other tissues, cardiac and skeletal muscle express two FXR1P isoforms that incorporate an additional exon-15. We report that recessive mutations in this particular exon of FXR1 cause congenital multi-minicore myopathy in humans and mice. Additionally, we show that while Myf5-dependent depletion of all FXR1P isoforms is neonatal lethal, mice carrying mutations in exon-15 display non-lethal myopathies which vary in severity depending on the specific effect of each mutation on the protein.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

