An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis
- PMID: 30770836
- PMCID: PMC6377607
- DOI: 10.1038/s41467-019-08783-0
An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis
Abstract
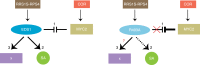
Plant intracellular NLR receptors recognise pathogen interference to trigger immunity but how NLRs signal is not known. Enhanced disease susceptibility1 (EDS1) heterodimers are recruited by Toll-interleukin1-receptor domain NLRs (TNLs) to transcriptionally mobilise resistance pathways. By interrogating the Arabidopsis EDS1 ɑ-helical EP-domain we identify positively charged residues lining a cavity that are essential for TNL immunity signalling, beyond heterodimer formation. Mutating a single, conserved surface arginine (R493) disables TNL immunity to an oomycete pathogen and to bacteria producing the virulence factor, coronatine. Plants expressing a weakly active EDS1R493A variant have delayed transcriptional reprogramming, with severe consequences for resistance and countering bacterial coronatine repression of early immunity genes. The same EP-domain surface is utilised by a non-TNL receptor RPS2 for bacterial immunity, indicating that the EDS1 EP-domain signals in resistance conferred by different NLR receptor types. These data provide a unique structural insight to early downstream signalling in NLR receptor immunity.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

