YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo
- PMID: 30773489
- PMCID: PMC6435425
- DOI: 10.1016/j.devcel.2019.01.017
YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo
Abstract
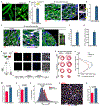
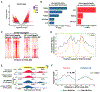
Specialized adult somatic cells, such as cardiomyocytes (CMs), are highly differentiated with poor renewal capacity, an integral reason underlying organ failure in disease and aging. Among the least renewable cells in the human body, CMs renew approximately 1% annually. Consistent with poor CM turnover, heart failure is the leading cause of death. Here, we show that an active version of the Hippo pathway effector YAP, termed YAP5SA, partially reprograms adult mouse CMs to a more fetal and proliferative state. One week after induction, 19% of CMs that enter S-phase do so twice, CM number increases by 40%, and YAP5SA lineage CMs couple to pre-existing CMs. Genomic studies showed that YAP5SA increases chromatin accessibility and expression of fetal genes, partially reprogramming long-lived somatic cells in vivo to a primitive, fetal-like, and proliferative state.
Keywords: Hippo pathway; Yap; cardiomyocyte proliferation; chromatin accessibility; heart development; heart failure; regeneration; reprogramming; transcription.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Prometheus Unbound in Ya(p) Heart.Dev Cell. 2019 Mar 25;48(6):741-742. doi: 10.1016/j.devcel.2019.03.007. Dev Cell. 2019. PMID: 30913402
References
-
- Ahuja P, Perriard E, Perriard JC, and Ehler E (2004). Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. Journal of cell science 117, 3295–3306. - PubMed
-
- Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, and Bergmann O (2015). No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell 163, 1026–1036. - PubMed
-
- Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, et al. (2015). Dynamics of Cell Generation and Turnover in the Human Heart. Cell 161, 1566–1575. - PubMed
-
- Bersell K, Arab S, Haring B, and Kuhn B (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL118761/HL/NHLBI NIH HHS/United States
- R41 HL129570/HL/NHLBI NIH HHS/United States
- U54 HD083092/HD/NICHD NIH HHS/United States
- R01 HL127717/HL/NHLBI NIH HHS/United States
- R01 HL091947/HL/NHLBI NIH HHS/United States
- UM1 HG006348/HG/NHGRI NIH HHS/United States
- U54 HG006348/HG/NHGRI NIH HHS/United States
- R01 HL130804/HL/NHLBI NIH HHS/United States
- R01 DE023177/DE/NIDCR NIH HHS/United States
- F31 HL136065/HL/NHLBI NIH HHS/United States
- R01 HL117641/HL/NHLBI NIH HHS/United States
- R01 AR061370/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

