Metabolite profiles reveal interspecific variation in operation of the Calvin-Benson cycle in both C4 and C3 plants
- PMID: 30773587
- PMCID: PMC6436152
- DOI: 10.1093/jxb/erz051
Metabolite profiles reveal interspecific variation in operation of the Calvin-Benson cycle in both C4 and C3 plants
Abstract
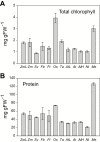
Low atmospheric CO2 in recent geological time led to the evolution of carbon-concentrating mechanisms (CCMs) such as C4 photosynthesis in >65 terrestrial plant lineages. We know little about the impact of low CO2 on the Calvin-Benson cycle (CBC) in C3 species that did not evolve CCMs, representing >90% of terrestrial plant species. Metabolite profiling provides a top-down strategy to investigate the operational balance in a pathway. We profiled CBC intermediates in a panel of C4 (Zea mays, Setaria viridis, Flaveria bidentis, and F. trinervia) and C3 species (Oryza sativa, Triticium aestivum, Arabidopsis thaliana, Nicotiana tabacum, and Manihot esculenta). Principal component analysis revealed differences between C4 and C3 species that were driven by many metabolites, including lower ribulose 1,5-bisphosphate in C4 species. Strikingly, there was also considerable variation between C3 species. This was partly due to different chlorophyll and protein contents, but mainly to differences in relative levels of metabolites. Correlation analysis indicated that one contributory factor was the balance between fructose-1,6-bisphosphatase, sedoheptulose-1,7-bisphosphatase, phosphoribulokinase, and Rubisco. Our results point to the CBC having experienced different evolutionary trajectories in C3 species since the ancestors of modern plant lineages diverged. They underline the need to understand CBC operation in a wide range of species.
Keywords: C3; C4; Calvin–Benson cycle; interspecies variation; metabolite profiles; photosynthesis.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures






Comment in
-
Variations in the Calvin-Benson cycle: selection pressures and optimization?J Exp Bot. 2019 Mar 27;70(6):1697-1701. doi: 10.1093/jxb/erz078. J Exp Bot. 2019. PMID: 30916343 Free PMC article.
References
-
- Adam NR. 2017. C3 carbon reduction cycle: eLS. Chichester: John Wiley & Sons, Ltd.
-
- Arrivault S, Guenther M, Fry SC, Fuenfgeld MM, Veyel D, Mettler-Altmann T, Stitt M, Lunn JE. 2015. Synthesis and use of stable-isotope-labeled internal standards for quantification of phosphorylated metabolites by LC-MS/MS. Analytical Chemistry 87, 6896–6904. - PubMed
-
- Arrivault S, Guenther M, Ivakov A, Feil R, Vosloh D, van Dongen JT, Sulpice R, Stitt M. 2009. Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. The Plant Journal 59, 824–839. - PubMed
-
- Awoyinka AF, Abegunde VO, Adewusi SR. 1995. Nutrient content of young cassava leaves and assessment of their acceptance as a green vegetable in Nigeria. Plant Foods for Human Nutrition 47, 21–28. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

