Teneurin C-Terminal Associated Peptide (TCAP)-1 and Latrophilin Interaction in HEK293 Cells: Evidence for Modulation of Intercellular Adhesion
- PMID: 30774623
- PMCID: PMC6367273
- DOI: 10.3389/fendo.2019.00022
Teneurin C-Terminal Associated Peptide (TCAP)-1 and Latrophilin Interaction in HEK293 Cells: Evidence for Modulation of Intercellular Adhesion
Abstract
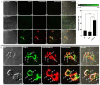
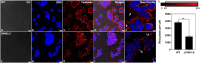
The teneurins are a family of four transmembrane proteins essential to intercellular adhesion processes, and are required for the development and maintenance of tissues. The Adhesion G protein-coupled receptor (GPCR) subclass latrophilins (ADGRL), or simply the latrophilins (LPHN), are putative receptors of the teneurins and act, in part, to mediate intercellular adhesion via binding with the teneurin extracellular region. At the distal tip of the extracellular region of each teneurin lies a peptide sequence termed the teneurin C-terminal associated peptide (TCAP). TCAP-1, associated with teneurin-1, is itself bioactive, suggesting that TCAP is a critical functional region of teneurin. However, the role of TCAP-1 has not been established with respect to its ability to interact with LPHN to induce downstream effects. To establish that TCAP-1 binds to LPHN1, a FLAG-tagged hormone binding domain (HBD) of LPHN1 and a GFP-tagged TCAP-1 peptide were co-expressed in HEK293 cells. Both immunoreactive epitopes were co-localized as a single band after immunoprecipitation, indicating an association between the two proteins. Moreover, fluorescent co-labeling occurred at the plasma membrane of LPHN1 over-expressing cells when treated with a FITC-tagged TCAP-1 variant. Expression of LPHN1 and treatment with TCAP-1 modulated the actin-based cytoskeleton in these cells in a manner consistent with previously reported actions of TCAP-1 and affected the overall morphology and aggregation of the cells. This study indicates that TCAP-1 may associate directly with LPHN1 and could play a role in the modulation of cytoskeletal organization and intercellular adhesion and aggregation via this interaction.
Keywords: GPCR; LPHN; TCAP; adhesion; latrophilin; peptides; receptor-ligand interaction; teneurin.
Figures






Similar articles
-
Role of Teneurin C-Terminal Associated Peptides (TCAP) on Intercellular Adhesion and Communication.Front Neurosci. 2022 May 2;16:868541. doi: 10.3389/fnins.2022.868541. eCollection 2022. Front Neurosci. 2022. PMID: 35585927 Free PMC article. Review.
-
Distal extracellular teneurin region (teneurin C-terminal associated peptide; TCAP) possesses independent intracellular calcium regulating actions, in vitro: A potential antagonist of corticotropin-releasing factor (CRF).Biochem Biophys Rep. 2022 Nov 26;32:101397. doi: 10.1016/j.bbrep.2022.101397. eCollection 2022 Dec. Biochem Biophys Rep. 2022. PMID: 36467544 Free PMC article.
-
Ancient interaction between the teneurin C-terminal associated peptides (TCAP) and latrophilin ligand-receptor coupling: a role in behavior.Front Neurosci. 2015 Apr 24;9:146. doi: 10.3389/fnins.2015.00146. eCollection 2015. Front Neurosci. 2015. PMID: 25964737 Free PMC article. Review.
-
Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing.J Biol Chem. 2014 Jan 3;289(1):387-402. doi: 10.1074/jbc.M113.504779. Epub 2013 Nov 22. J Biol Chem. 2014. PMID: 24273166 Free PMC article.
-
C-terminal processing of the teneurin proteins: independent actions of a teneurin C-terminal associated peptide in hippocampal cells.Mol Cell Neurosci. 2013 Jan;52:38-50. doi: 10.1016/j.mcn.2012.09.006. Epub 2012 Sep 28. Mol Cell Neurosci. 2013. PMID: 23026563
Cited by
-
Synthetic Peptides as Therapeutic Agents: Lessons Learned From Evolutionary Ancient Peptides and Their Transit Across Blood-Brain Barriers.Front Endocrinol (Lausanne). 2019 Nov 12;10:730. doi: 10.3389/fendo.2019.00730. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 31781029 Free PMC article.
-
Role of Teneurin C-Terminal Associated Peptides (TCAP) on Intercellular Adhesion and Communication.Front Neurosci. 2022 May 2;16:868541. doi: 10.3389/fnins.2022.868541. eCollection 2022. Front Neurosci. 2022. PMID: 35585927 Free PMC article. Review.
-
A novel role for Teneurin C-terminal Associated Peptide (TCAP) in the regulation of cardiac activity in the Sydney rock oyster, Saccostrea glomerata.Front Endocrinol (Lausanne). 2023 Feb 6;14:1020368. doi: 10.3389/fendo.2023.1020368. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36814576 Free PMC article.
-
Signaling Pathways Regulating the Expression of the Glioblastoma Invasion Factor TENM1.Biomedicines. 2022 May 10;10(5):1104. doi: 10.3390/biomedicines10051104. Biomedicines. 2022. PMID: 35625843 Free PMC article. Review.
-
Distal extracellular teneurin region (teneurin C-terminal associated peptide; TCAP) possesses independent intracellular calcium regulating actions, in vitro: A potential antagonist of corticotropin-releasing factor (CRF).Biochem Biophys Rep. 2022 Nov 26;32:101397. doi: 10.1016/j.bbrep.2022.101397. eCollection 2022 Dec. Biochem Biophys Rep. 2022. PMID: 36467544 Free PMC article.
References
-
- Nunes SM, Ferralli J, Choi K, Brown-Luedi M, Minet AD, Chiquet-Ehrismann R. The intracellular domain of teneurin-1 interacts with MBD1 and CAP/ponsin resulting in subcellular codistribution and translocation to the nuclear matrix. Exp Cell Res. (2005) 305:122–32. 10.1016/j.yexcr.2004.12.020 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

