A synthetic approach to 'click' neoglycoprotein analogues of EPO employing one-pot native chemical ligation and CuAAC chemistry
- PMID: 30774876
- PMCID: PMC6345360
- DOI: 10.1039/c8sc03409e
A synthetic approach to 'click' neoglycoprotein analogues of EPO employing one-pot native chemical ligation and CuAAC chemistry
Abstract
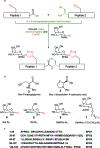
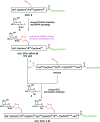
The clinical significance of batch-wise variability on the pharmacokinetics and potency of commercial erythropoietin (EPO), prepared recombinantly as a heterogeneous mixture of glycoforms, necessitates the development of synthetic strategies to afford homogenous EPO formulations. Herein we present a previously unexplored and divergent route towards 'click' neoglycoprotein analogues of EPO, employing one-pot native chemical ligation (NCL) of alkynylated peptides and copper-catalysed azide-alkyne cycloaddition (CuAAC) with azido monosaccharides. By design, our synthetic platform permits glycosylation at virtually any stage, providing flexibility for the synthesis of various glycoforms for biological analysis. Insights obtained from attempted folding of our 'click' neoglycoprotein EPO analogue, bearing four different neutral sugar moieties, highlight the important role played by the charged oligosaccharides present in native EPO glycoproteins.
Figures






References
-
- Graber S. E., Krantz S. Annu. Rev. Med. 1978;29:51–66. - PubMed
-
- Eschbach J. W., Egrie J. C., Downing M. R., Browne J. K., Adamson J. W. N. Engl. J. Med. 1987;316:73–78. - PubMed
-
- Bohlius J., Wilson J., Seidenfeld J., Piper M., Schwarzer G., Sandercock J., Trelle S., Weingart O., Bayliss S., Djulbegovic B. J. Natl. Cancer Inst. 2006;98:708–714. - PubMed
-
- Recny M. A., Scoble H., Kim Y. J. Biol. Chem. 1987;262:17156–17163. - PubMed
-
- Sasaki H., Bothner B., Dell A., Fukuda M. J. Biol. Chem. 1987;262:12059–12076. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

