Syntheses of defined sulfated oligohyaluronans reveal structural effects, diversity and thermodynamics of GAG-protein binding
- PMID: 30774881
- PMCID: PMC6346292
- DOI: 10.1039/c8sc03649g
Syntheses of defined sulfated oligohyaluronans reveal structural effects, diversity and thermodynamics of GAG-protein binding
Abstract
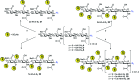
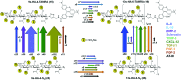
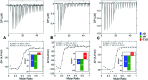
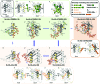
Binding of sulfated glycosaminoglycans (GAG) to a wide spectrum of extracellular regulatory proteins is crucial for physiological processes such as cell growth, migration, tissue homeostasis and repair. Thus, GAG derivatives exhibit great relevance in the development of innovative biomaterials for tissue regeneration therapies. We present a synthetic strategy for the preparation of libraries of defined sulfated oligohyaluronans as model GAG systematically varied in length, sulfation pattern and anomeric substitution in order to elucidate the effects of these parameters on GAG recognition by regulatory proteins. Through an experimental and computational approach using fluorescence polarization, ITC, docking and molecular dynamics simulations we investigate the binding of these functionalized GAG derivatives to ten representative regulatory proteins including IL-8, IL-10, BMP-2, sclerostin, TIMP-3, CXCL-12, TGF-β, FGF-1, FGF-2, and AT-III, and we establish structure-activity relationships for GAG recognition. Binding is mainly driven by enthalpy with only minor entropic contributions. In several cases binding is determined by GAG length, and in all cases by the position and number of sulfates. Affinities strongly depend on the anomeric modification of the GAG. Highest binding affinities are effected by anomeric functionalization with large fluorophores and by GAG dimerization. Our experimental and theoretical results suggest that the diversity of GAG binding sites and modes is responsible for the observed high affinities and other binding features. The presented new insights into GAG-protein recognition will be of relevance to guide the design of GAG derivatives with customized functions for the engineering of new biomaterials.
Figures

References
-
- Gandhi N. S., Mancera R. L. Chem. Biol. Drug Des. 2008;72:455–482. - PubMed
-
- Raman R., Sasisekharan V., Sasisekharan R. Chem. Biol. 2005;12:267–277. - PubMed
- Mizumoto S., Yamada S., Sugahara K. Curr. Opin. Struct. Biol. 2015;34:35–42. - PubMed
- Esko J. D. and Linhardt R. J., in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart and M. E. Etzler, Cold Spring Harbor Laboratory Press The Consortium of Glycobiology Editors, La Jolla, California, Cold Spring Harbor, NY, 2009. - PubMed
-
- Habuchi H., Habuchi O., Kimata K. Glycoconjugate J. 2004;21:47–52. - PubMed
- Martinez P., Denys A., Delos M., Sikora A. S., Carpentier M., Julien S., Pestel J., Allain F. Glycobiology. 2015;25:502–513. - PubMed
- Zong C., Venot A., Li X., Lu W., Xiao W., Wilkes J. L., Salanga C. L., Handel T. M., Wang L., Wolfert M. A., Boons G. J. J. Am. Chem. Soc. 2017;139:9534–9543. - PMC - PubMed
- Zong C., Huang R., Condac E., Chiu Y., Xiao W., Li X., Lu W., Ishihara M., Wang S., Ramiah A., Stickney M., Azadi P., Amster I. J., Moremen K. W., Wang L., Sharp J. S., Boons G. J. J. Am. Chem. Soc. 2016;138:13059–13067. - PMC - PubMed
- Gama C. I., Tully S. E., Sotogaku N., Clark P. M., Rawat M., Vaidehi N., Goddard III W. A., Nishi A., Hsieh-Wilson L. C. Nat. Chem. Biol. 2006;2:467–473. - PubMed
- Angulo J., Ojeda R., de Paz J. L., Lucas R., Nieto P. M., Lozano R. M., Redondo-Horcajo M., Gimenez-Gallego G., Martin-Lomas M. ChemBioChem. 2004;5:55–61. - PubMed
- Raman K., Mencio C., Desai U. R., Kuberan B. Mol. Pharm. 2013;10:1442–1449. - PMC - PubMed
- Sankarayanarayanan N. V., Strebel T. R., Boothello R. S., Sheerin K., Raghuraman A., Sallas F., Mosier P. D., Watermeyer N. D., Oscarson S., Desai U. R. Angew. Chem., Int. Ed. 2017;56:2312–2317. - PMC - PubMed
-
- Gandhi N. S., Mancera R. L. Glycobiology. 2009;19:1103–1115. - PubMed
- Samsonov S. A., Teyra J., Pisabarro M. T. J. Comput.-Aided Mol. Des. 2011;25:477–489. - PMC - PubMed
- Sarkar A., Desai U. R. PLoS One. 2015;10:e0141127. - PMC - PubMed
- Sarkar A., Yu W., Desai U. R., MacKerell A. D., Mosier P. D. Glycobiology. 2016;26:1041–1047. - PMC - PubMed
-
- Salbach J., Rachner T. D., Rauner M., Hempel U., Anderegg U., Franz S., Simon J. C., Hofbauer L. C. J. Mol. Med. 2012;90:625–635. - PubMed
- Martin P. Science. 1997;276:75–81. - PubMed
- Picke A. K., Salbach-Hirsch J., Hintze V., Rother S., Rauner M., Kascholke C., Moller S., Bernhardt R., Rammelt S., Pisabarro M. T., Ruiz-Gomez G., Schnabelrauch M., Schulz-Siegmund M., Hacker M. C., Scharnweber D., Hofbauer C., Hofbauer L. C. Biomaterials. 2016;96:11–23. - PubMed
- Kuenze G., Koehling S., Vogel A., Rademann J., Huster D. J. Biol. Chem. 2016;291:3100–3113. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

