Inhibition of histone methyltransferase DOT1L silences ERα gene and blocks proliferation of antiestrogen-resistant breast cancer cells
- PMID: 30775443
- PMCID: PMC6365116
- DOI: 10.1126/sciadv.aav5590
Inhibition of histone methyltransferase DOT1L silences ERα gene and blocks proliferation of antiestrogen-resistant breast cancer cells
Abstract
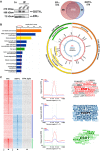
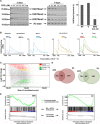
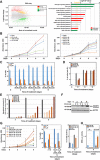
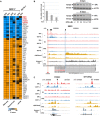
Breast cancer (BC) resistance to endocrine therapy results from constitutively active or aberrant estrogen receptor α (ERα) signaling, and ways to block ERα pathway in these tumors are sought after. We identified the H3K79 methyltransferase DOT1L as a novel cofactor of ERα in BC cell chromatin, where the two proteins colocalize to regulate estrogen target gene transcription. DOT1L blockade reduces proliferation of hormone-responsive BC cells in vivo and in vitro, consequent to cell cycle arrest and apoptotic cell death, with widespread effects on ER-dependent gene transcription, including ERα and FOXA1 gene silencing. Antiestrogen-resistant BC cells respond to DOT1L inhibition also in mouse xenografts, with reduction in ERα levels, H3K79 methylation, and tumor growth. These results indicate that DOT1L is an exploitable epigenetic target for treatment of endocrine therapy-resistant ERα-positive BCs.
Figures


References
-
- Tryfonidis K., Zardavas D., Katzenellenbogen B. S., Piccart M., Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treat. Rev. 50, 68–81 (2016). - PubMed
-
- Bailey S. D., Desai K., Kron K. J., Mazrooei P., Sinnott-Armstrong N. A., Treloar A. E., Dowar M., Thu K. L., Cescon D. W., Silvester J., Yang S. Y., Wu X., Pezo R. C., Haibe-Kains B., Mak T. W., Bedard P. L., Pugh T. J., Sallari R. C., Lupien M., Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat. Genet. 48, 1260–1266 (2016). - PMC - PubMed
-
- Razavi P., Chang M. T., Xu G., Bandlamudi C., Ross D. S., Vasan N., Cai Y., Bielski C. M., Donoghue M. T. A., Jonsson P., Penson A., Shen R., Pareja F., Kundra R., Middha S., Cheng M. L., Zehir A., Kandoth C., Patel R., Huberman K., Smyth L. M., Jhaveri K., Modi S., Traina T. A., Dang C., Zhang W., Weigelt B., Li B. T., Ladanyi M., Hyman D. M., Schultz N., Robson M. E., Hudis C., Brogi E., Viale A., Norton L., Dickler M. N., Berger M. F., Iacobuzio-Donahue C. A., Chandarlapaty S., Scaltriti M., Reis-Filho J. S., Solit D. B., Taylor B. S., Baselga J., The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34, 427–438.e6 (2018). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

