Relative Binding Affinity Prediction of Charge-Changing Sequence Mutations with FEP in Protein-Protein Interfaces
- PMID: 30776430
- PMCID: PMC6453258
- DOI: 10.1016/j.jmb.2019.02.003
Relative Binding Affinity Prediction of Charge-Changing Sequence Mutations with FEP in Protein-Protein Interfaces
Abstract
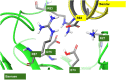
Building on the substantial progress that has been made in using free energy perturbation (FEP) methods to predict the relative binding affinities of small molecule ligands to proteins, we have previously shown that results of similar quality can be obtained in predicting the effect of mutations on the binding affinity of protein-protein complexes. However, these results were restricted to mutations which did not change the net charge of the side chains due to known difficulties with modeling perturbations involving a change in charge in FEP. Various methods have been proposed to address this problem. Here we apply the co-alchemical water approach to study the efficacy of FEP calculations of charge changing mutations at the protein-protein interface for the antibody-gp120 system investigated previously and three additional complexes. We achieve an overall root mean square error of 1.2 kcal/mol on a set of 106 cases involving a change in net charge selected by a simple suitability filter using side-chain predictions and solvent accessible surface area to be relevant to a biologic optimization project. Reasonable, although less precise, results are also obtained for the 44 more challenging mutations that involve buried residues, which may in some cases require substantial reorganization of the local protein structure, which can extend beyond the scope of a typical FEP simulation. We believe that the proposed prediction protocol will be of sufficient efficiency and accuracy to guide protein engineering projects for which optimization and/or maintenance of a high degree of binding affinity is a key objective.
Keywords: antibodies; free energy perturbation; protein-protein binding.
Copyright © 2019 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



Similar articles
-
Free Energy Perturbation Calculation of Relative Binding Free Energy between Broadly Neutralizing Antibodies and the gp120 Glycoprotein of HIV-1.J Mol Biol. 2017 Apr 7;429(7):930-947. doi: 10.1016/j.jmb.2016.11.021. Epub 2016 Nov 28. J Mol Biol. 2017. PMID: 27908641 Free PMC article.
-
Protein-Ligand Binding Free Energy Calculations with FEP.Methods Mol Biol. 2019;2022:201-232. doi: 10.1007/978-1-4939-9608-7_9. Methods Mol Biol. 2019. PMID: 31396905
-
Improving the Accuracy of Protein Thermostability Predictions for Single Point Mutations.Biophys J. 2020 Jul 7;119(1):115-127. doi: 10.1016/j.bpj.2020.05.020. Epub 2020 May 29. Biophys J. 2020. PMID: 32533939 Free PMC article.
-
Free Energy Calculations for Protein-Ligand Binding Prediction.Methods Mol Biol. 2021;2266:203-226. doi: 10.1007/978-1-0716-1209-5_12. Methods Mol Biol. 2021. PMID: 33759129 Review.
-
Structural studies of human HIV-1 V3 antibodies.Hum Antibodies. 2005;14(3-4):73-80. Hum Antibodies. 2005. PMID: 16720977 Review.
Cited by
-
High-Throughput Molecular Dynamics-Based Alchemical Free Energy Calculations for Predicting the Binding Free Energy Change Associated with the Selected Omicron Mutations in the Spike Receptor-Binding Domain of SARS-CoV-2.Biomedicines. 2022 Nov 1;10(11):2779. doi: 10.3390/biomedicines10112779. Biomedicines. 2022. PMID: 36359299 Free PMC article.
-
Predicting the mutation effects of protein-ligand interactions via end-point binding free energy calculations: strategies and analyses.J Cheminform. 2022 Aug 20;14(1):56. doi: 10.1186/s13321-022-00639-y. J Cheminform. 2022. PMID: 35987841 Free PMC article.
-
On the Rapid Calculation of Binding Affinities for Antigen and Antibody Design and Affinity Maturation Simulations.Antibodies (Basel). 2022 Aug 3;11(3):51. doi: 10.3390/antib11030051. Antibodies (Basel). 2022. PMID: 35997345 Free PMC article.
-
Alkaloids in Contemporary Drug Discovery to Meet Global Disease Needs.Molecules. 2021 Jun 22;26(13):3800. doi: 10.3390/molecules26133800. Molecules. 2021. PMID: 34206470 Free PMC article. Review.
-
Identification of SARS-CoV-2 Main Protease Inhibitors Using Structure Based Virtual Screening and Molecular Dynamics Simulation of DrugBank Database.ChemistrySelect. 2021 May 27;6(20):4991-5013. doi: 10.1002/slct.202100854. Epub 2021 Jun 18. ChemistrySelect. 2021. PMID: 34541295 Free PMC article.
References
-
- Stamatatos L., Morris L., Burton D.R., Mascola J.R. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 2009;15:866–870. - PubMed
-
- Pawson T., Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;vol. 300(80):445 LP–452. http://science.sciencemag.org/content/300/5618/445.abstract - PubMed
-
- Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;2836:369–387. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

