Morphologic determinant of tight junctions revealed by claudin-3 structures
- PMID: 30778075
- PMCID: PMC6379431
- DOI: 10.1038/s41467-019-08760-7
Morphologic determinant of tight junctions revealed by claudin-3 structures
Abstract
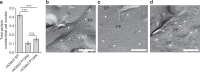
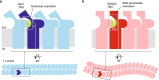
Tight junction is a cell adhesion apparatus functioning as barrier and/or channel in the paracellular spaces of epithelia. Claudin is the major component of tight junction and polymerizes to form tight junction strands with various morphologies that may correlate with their functions. Here we present the crystal structure of mammalian claudin-3 at 3.6 Å resolution. The third transmembrane helix of claudin-3 is clearly bent compared with that of other subtypes. Structural analysis of additional two mutants with a single mutation representing other subtypes in the third helix indicates that this helix takes a bent or straight structure depending on the residue. The presence or absence of the helix bending changes the positions of residues related to claudin-claudin interactions and affects the morphology and adhesiveness of the tight junction strands. These results evoke a model for tight junction strand formation with different morphologies - straight or curvy strands - observed in native epithelia.
Conflict of interest statement
The authors declare no competing interests.
Figures

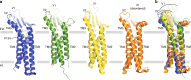


Similar articles
-
Polar and charged extracellular residues conserved among barrier-forming claudins contribute to tight junction strand formation.Ann N Y Acad Sci. 2017 Jun;1397(1):143-156. doi: 10.1111/nyas.13341. Epub 2017 Apr 17. Ann N Y Acad Sci. 2017. PMID: 28415153
-
Assembly of Tight Junction Strands: Claudin-10b and Claudin-3 Form Homo-Tetrameric Building Blocks that Polymerise in a Channel-Independent Manner.J Mol Biol. 2020 Mar 27;432(7):2405-2427. doi: 10.1016/j.jmb.2020.02.034. Epub 2020 Mar 4. J Mol Biol. 2020. PMID: 32142789
-
Enhancement of the thermostability of mouse claudin-3 on complex formation with the carboxyl-terminal region of Clostridium perfringens enterotoxin improves crystal quality.Acta Crystallogr F Struct Biol Commun. 2018 Mar 1;74(Pt 3):150-155. doi: 10.1107/S2053230X18002005. Epub 2018 Feb 26. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 29497018 Free PMC article.
-
Computational Models of Claudin Assembly in Tight Junctions and Strand Properties.Int J Mol Sci. 2024 Mar 16;25(6):3364. doi: 10.3390/ijms25063364. Int J Mol Sci. 2024. PMID: 38542338 Free PMC article. Review.
-
Tight junctions of the proximal tubule and their channel proteins.Pflugers Arch. 2017 Aug;469(7-8):877-887. doi: 10.1007/s00424-017-2001-3. Epub 2017 Jun 9. Pflugers Arch. 2017. PMID: 28600680 Review.
Cited by
-
Cell adhesion signals regulate the nuclear receptor activity.Proc Natl Acad Sci U S A. 2019 Dec 3;116(49):24600-24609. doi: 10.1073/pnas.1913346116. Epub 2019 Nov 18. Proc Natl Acad Sci U S A. 2019. PMID: 31740618 Free PMC article.
-
Overall protein structure quality assessment using hydrogen-bonding parameters.Acta Crystallogr D Struct Biol. 2023 Aug 1;79(Pt 8):684-693. doi: 10.1107/S2059798323005077. Epub 2023 Jul 11. Acta Crystallogr D Struct Biol. 2023. PMID: 37431759 Free PMC article.
-
Single-Blinded Study Highlighting the Differences between the Small Intestines of Neonatal and Weaned Piglets.Animals (Basel). 2021 Jan 21;11(2):271. doi: 10.3390/ani11020271. Animals (Basel). 2021. PMID: 33494523 Free PMC article.
-
MTCH2 promotes the malignant progression of ovarian cancer through the upregulation of AIMP2 expression levels, mitochondrial dysfunction and by mediating energy metabolism.Oncol Lett. 2024 Aug 12;28(4):492. doi: 10.3892/ol.2024.14625. eCollection 2024 Oct. Oncol Lett. 2024. PMID: 39185493 Free PMC article.
-
AMPA receptor structure and auxiliary subunits.J Physiol. 2021 Jan;599(2):453-469. doi: 10.1113/JP278701. Epub 2020 Feb 18. J Physiol. 2021. PMID: 32004381 Free PMC article. Review.
References
-
- Staehelin LA. Further observations on the fine structure of freeze-cleaved tight junctions. J. Cell. Sci. 1973;13:763–786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

