The effect of Robertsonian translocations on the intranuclear positioning of NORs (nucleolar organizing regions) in human sperm cells
- PMID: 30778082
- PMCID: PMC6379386
- DOI: 10.1038/s41598-019-38478-x
The effect of Robertsonian translocations on the intranuclear positioning of NORs (nucleolar organizing regions) in human sperm cells
Abstract
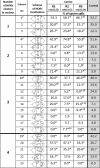
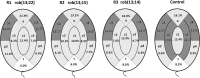
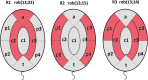
Only a few studies have described sperm chromosome intranuclear positioning changes in men with reproductive failure and an incorrect somatic karyotype. We studied the influence of Robertsonian translocations on the acrocentric chromosome positioning in human sperm cells. The basis of the analysis was the localization of NORs (nucleolar organizing regions) in sperm nuclei from three Robertsonian translocation carriers, namely, rob(13;22), rob(13;15) and rob(13;14), with a known meiotic segregation pattern. All three carriers presented with a similar percentage of genetically normal sperm cells (i.e., approximately 40%). To visualize NORs, we performed 2D-FISH with directly labelled probes. We used the linear and radial topologies of the nucleus to analyse the NORs distribution. We found an affected positioning of NORs in each case of the Robertsonian translocations. Moreover, the NORs tended to group, most often in two clusters. Both in Robertsonian carriers and control sperm cells, NORs mostly colocalized in the medial areas of the nuclei. In the case of the Roberstonian carriers, NORs were mostly concentrated in the peripheral part of the medial area, in contrast to control sperm cells in which the distribution was more dispersed towards the internal area.
Conflict of interest statement
The authors declare no competing interests.
Figures
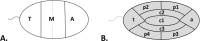
Similar articles
-
How much, if anything, do we know about sperm chromosomes of Robertsonian translocation carriers?Cell Mol Life Sci. 2020 Dec;77(23):4765-4785. doi: 10.1007/s00018-020-03560-5. Epub 2020 Jun 8. Cell Mol Life Sci. 2020. PMID: 32514588 Free PMC article. Review.
-
Meiotic and sperm aneuploidy studies in three carriers of Robertsonian translocations and small supernumerary marker chromosomes.Fertil Steril. 2015 May;103(5):1162-9.e7. doi: 10.1016/j.fertnstert.2015.02.006. Epub 2015 Mar 18. Fertil Steril. 2015. PMID: 25796321
-
Meiotic segregation analysis in male translocation carriers by using fluorescent in situ hybridization.Int J Androl. 2008 Feb;31(1):60-6. doi: 10.1111/j.1365-2605.2007.00759.x. Epub 2007 Apr 24. Int J Androl. 2008. PMID: 17459123
-
Meiotic segregation of rare Robertsonian translocations: sperm analysis of three t(14q;22q) cases.Hum Reprod. 2006 May;21(5):1166-71. doi: 10.1093/humrep/dei477. Epub 2006 Jan 26. Hum Reprod. 2006. PMID: 16439506
-
Meiotic segregation of translocations during male gametogenesis.Int J Androl. 2004 Aug;27(4):200-12. doi: 10.1111/j.1365-2605.2004.00490.x. Int J Androl. 2004. PMID: 15271199 Review.
Cited by
-
How much, if anything, do we know about sperm chromosomes of Robertsonian translocation carriers?Cell Mol Life Sci. 2020 Dec;77(23):4765-4785. doi: 10.1007/s00018-020-03560-5. Epub 2020 Jun 8. Cell Mol Life Sci. 2020. PMID: 32514588 Free PMC article. Review.
-
Prevalence and Phenotypic Impact of Robertsonian Translocations.Mol Syndromol. 2021 Mar;12(1):1-11. doi: 10.1159/000512676. Epub 2021 Feb 17. Mol Syndromol. 2021. PMID: 33776621 Free PMC article. Review.
References
-
- Robertson, W. R. B. Chromosome studies. I. Taxonomic relationships shown in the chromosomes of Tettigidae and Acrididae. V-shaped chromosomes and their significance in Acrididae, Locustidae and Gryllidae: chromosome and variation. J. Morph. 27, 179-331 (1916).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

