The DNA modification N6-methyl-2'-deoxyadenosine (m6dA) drives activity-induced gene expression and is required for fear extinction
- PMID: 30778148
- PMCID: PMC6462436
- DOI: 10.1038/s41593-019-0339-x
The DNA modification N6-methyl-2'-deoxyadenosine (m6dA) drives activity-induced gene expression and is required for fear extinction
Abstract
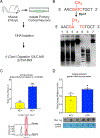
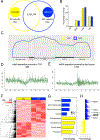
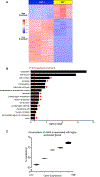
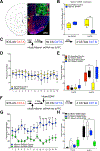
DNA modification is known to regulate experience-dependent gene expression. However, beyond cytosine methylation and its oxidated derivatives, very little is known about the functional importance of chemical modifications on other nucleobases in the brain. Here we report that in adult mice trained in fear extinction, the DNA modification N6-methyl-2'-deoxyadenosine (m6dA) accumulates along promoters and coding sequences in activated prefrontal cortical neurons. The deposition of m6dA is associated with increased genome-wide occupancy of the mammalian m6dA methyltransferase, N6amt1, and this correlates with extinction-induced gene expression. The accumulation of m6dA is associated with transcriptional activation at the brain-derived neurotrophic factor (Bdnf) P4 promoter, which is required for Bdnf exon IV messenger RNA expression and for the extinction of conditioned fear. These results expand the scope of DNA modifications in the adult brain and highlight changes in m6dA as an epigenetic mechanism associated with activity-induced gene expression and the formation of fear extinction memory.
Conflict of interest statement
Competing Interests:
The authors declare no competing financial interests.
Figures



References
-
- Baker-Andresen D, Ratnu VS & Bredy TW Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends in Neurosciences 36, 3–13 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

