Opposing roles of TCF7/LEF1 and TCF7L2 in cyclin D2 and Bmp4 expression and cardiomyocyte cell cycle control during late heart development
- PMID: 30778164
- PMCID: PMC6570565
- DOI: 10.1038/s41374-019-0204-2
Opposing roles of TCF7/LEF1 and TCF7L2 in cyclin D2 and Bmp4 expression and cardiomyocyte cell cycle control during late heart development
Abstract
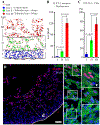
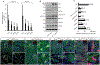
Bone morphogenetic protein (BMP) and Wnt pathways regulate cell proliferation and differentiation, but how these two pathways interact and mediate their nuclear actions in the heart, especially during late cardiac development, remains poorly defined. T-cell factor (TCF) and lymphoid enhancer factor (LEF) family transcriptional factors, including Lef1, Tcf7, Tcf7l1, and Tcf7l2, are important nuclear mediators of canonical Wnt/β-catenin signaling throughout cardiac development. We reveal that these TCF/LEF family members direct heart maturation through distinct temporal and spatial control. TCF7 and LEF1 decrease while TCF7L1 and TCF7L2 remain relatively stable during heart development. LEF1 is mainly expressed in mesenchymal cells in valvular regions. TCF7 and TCF7L1 are detected in the nucleus of mesothelial and endothelial cells, but not in cardiomyocytes or mesenchymal cells. Tcf7l2 is the primary TCF/LEF family member in cardiomyocytes and undergoes alternative splicing during heart development. A TCF7L2 intensity gradient opposite to that of β-catenin and cardiomyocyte proliferative activity is present in fetal hearts. Wnt activation by cardiac deletion of APC, a negative Wnt regulator, dramatically increases Cyclin D2 and Bmp4 expression. BMP signal transducing transcription factors, the mothers against decapentaplegic homologs (SMADs) are increasingly phosphorylated upon Wnt activation. LEF1/TCF7 displaces TCF7L2 and cooperates with pSMAD 1/5/8 in the regulatory elements of Cyclin D2 and Bmp4 promoters to promote β-catenin recruitment and transcriptional activation. Finally, we demonstrate that TCF7L2 is a transcriptional suppressor of Cyclin D2 and Bmp 4 in a cardiac cell line by overexpression and knockdown experiments.
Conflict of interest statement
Figures



Similar articles
-
Regulation of the TMEPAI promoter by TCF7L2: the C-terminal tail of TCF7L2 is essential to activate the TMEPAI gene.J Biochem. 2016 Jan;159(1):27-30. doi: 10.1093/jb/mvv117. Epub 2015 Nov 20. J Biochem. 2016. PMID: 26590303 Free PMC article.
-
Cell-context dependent TCF/LEF expression and function: alternative tales of repression, de-repression and activation potentials.Crit Rev Eukaryot Gene Expr. 2011;21(3):207-36. doi: 10.1615/critreveukargeneexpr.v21.i3.10. Crit Rev Eukaryot Gene Expr. 2011. PMID: 22111711 Free PMC article. Review.
-
Homeobox protein MSX-1 inhibits expression of bone morphogenetic protein 2, bone morphogenetic protein 4, and lymphoid enhancer-binding factor 1 via Wnt/β-catenin signaling to prevent differentiation of dental mesenchymal cells during the late bell stage.Eur J Oral Sci. 2018 Feb;126(1):1-12. doi: 10.1111/eos.12390. Epub 2017 Nov 17. Eur J Oral Sci. 2018. PMID: 29148101
-
A β-catenin-driven switch in TCF/LEF transcription factor binding to DNA target sites promotes commitment of mammalian nephron progenitor cells.Elife. 2021 Feb 15;10:e64444. doi: 10.7554/eLife.64444. Elife. 2021. PMID: 33587034 Free PMC article.
-
TCF/LEF Transcription Factors: An Update from the Internet Resources.Cancers (Basel). 2016 Jul 20;8(7):70. doi: 10.3390/cancers8070070. Cancers (Basel). 2016. PMID: 27447672 Free PMC article. Review.
Cited by
-
Characterizing the SREB G protein-coupled receptor family in fish: Brain gene expression and genomic differences in upstream transcription factor binding sites.Comp Biochem Physiol A Mol Integr Physiol. 2023 Nov;285:111507. doi: 10.1016/j.cbpa.2023.111507. Epub 2023 Aug 21. Comp Biochem Physiol A Mol Integr Physiol. 2023. PMID: 37611891 Free PMC article.
-
Cardiomyocyte Death and Genome-Edited Stem Cell Therapy for Ischemic Heart Disease.Stem Cell Rev Rep. 2021 Aug;17(4):1264-1279. doi: 10.1007/s12015-020-10096-5. Epub 2021 Jan 25. Stem Cell Rev Rep. 2021. PMID: 33492627 Free PMC article. Review.
-
Expression dynamics of HAND1/2 in in vitro human cardiomyocyte differentiation.Stem Cell Reports. 2021 Aug 10;16(8):1906-1922. doi: 10.1016/j.stemcr.2021.06.014. Epub 2021 Jul 22. Stem Cell Reports. 2021. PMID: 34297940 Free PMC article.
-
Metastasising ameloblastoma or ameloblastic carcinoma? A case report with mutation analyses.BMC Oral Health. 2023 Aug 12;23(1):563. doi: 10.1186/s12903-023-03259-6. BMC Oral Health. 2023. PMID: 37573343 Free PMC article. Review.
-
Genome-Wide Assessment of DNA Methylation in Chicken Cardiac Tissue Exposed to Different Incubation Temperatures and CO2 Levels.Front Genet. 2020 Oct 28;11:558189. doi: 10.3389/fgene.2020.558189. eCollection 2020. Front Genet. 2020. PMID: 33193638 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

