Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development
- PMID: 30778237
- PMCID: PMC6402993
- DOI: 10.1038/s41594-019-0189-y
Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development
Abstract
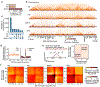
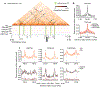
Germ cells manifest a unique gene expression program and regain totipotency in the zygote. Here, we perform Hi-C analysis to examine 3D chromatin organization in male germ cells during spermatogenesis. We show that the highly compartmentalized 3D chromatin organization characteristic of interphase nuclei is attenuated in meiotic prophase. Meiotic prophase is predominated by short-range intrachromosomal interactions that represent a condensed form akin to that of mitotic chromosomes. However, unlike mitotic chromosomes, meiotic chromosomes display weak genomic compartmentalization, weak topologically associating domains, and localized point interactions in prophase. In postmeiotic round spermatids, genomic compartmentalization increases and gives rise to the strong compartmentalization seen in mature sperm. The X chromosome lacks domain organization during meiotic sex-chromosome inactivation. We propose that male meiosis occurs amid global reprogramming of 3D chromatin organization and that strengthening of chromatin compartmentalization takes place in spermiogenesis to prepare the next generation of life.
Figures

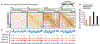


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

