Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome
- PMID: 30778239
- PMCID: PMC6546610
- DOI: 10.1038/s41591-018-0338-6
Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome
Abstract
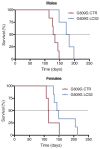
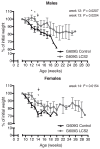
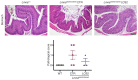
CRISPR/Cas9-based therapies hold considerable promise for the treatment of genetic diseases. Among these, Hutchinson-Gilford progeria syndrome, caused by a point mutation in the LMNA gene, stands out as a potential candidate. Here, we explore the efficacy of a CRISPR/Cas9-based approach that reverts several alterations in Hutchinson-Gilford progeria syndrome cells and mice by introducing frameshift mutations in the LMNA gene.
Conflict of interest statement
Figures
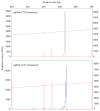
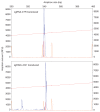
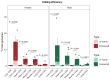
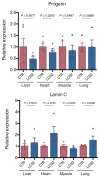




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

