The variable quality of metadata about biological samples used in biomedical experiments
- PMID: 30778255
- PMCID: PMC6380228
- DOI: 10.1038/sdata.2019.21
The variable quality of metadata about biological samples used in biomedical experiments
Abstract
We present an analytical study of the quality of metadata about samples used in biomedical experiments. The metadata under analysis are stored in two well-known databases: BioSample-a repository managed by the National Center for Biotechnology Information (NCBI), and BioSamples-a repository managed by the European Bioinformatics Institute (EBI). We tested whether 11.4 M sample metadata records in the two repositories are populated with values that fulfill the stated requirements for such values. Our study revealed multiple anomalies in the metadata. Most metadata field names and their values are not standardized or controlled. Even simple binary or numeric fields are often populated with inadequate values of different data types. By clustering metadata field names, we discovered there are often many distinct ways to represent the same aspect of a sample. Overall, the metadata we analyzed reveal that there is a lack of principled mechanisms to enforce and validate metadata requirements. The significant aberrancies that we found in the metadata are likely to impede search and secondary use of the associated datasets.
Conflict of interest statement
The authors declare no competing interests.
Figures
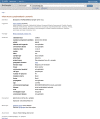

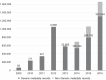




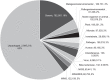

References
-
- Bruce T. R., Hillmann D. I. The Continuum of Metadata Quality: Defining, Expressing, Exploiting. in Metadata in Practice, (eds Hillmann D. I. & Westbrooks E. L.) 238–256 (ALA Editions, 2004).
-
- Park J.-R. Metadata Quality in Digital Repositories: A Survey of the Current State of the Art. Cataloging & Classification Quarterly 47, 213–228 (2009).
-
- Park J.-R. & Tosaka Y. Metadata Quality Control in Digital Repositories and Collections: Criteria, Semantics, and Mechanisms. Cataloging & Classification Quarterly 48, 696–715 (2010).
-
- Zaveri A. & Dumontier M. MetaCrowd: Crowdsourcing Biomedical Metadata Quality Assessment. in Proceedings of the Bio-Ontologies Workshop (2017).
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

