An ancillary genomics system to support the return of pharmacogenomic results
- PMID: 30778576
- PMCID: PMC6402311
- DOI: 10.1093/jamia/ocy187
An ancillary genomics system to support the return of pharmacogenomic results
Abstract
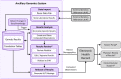
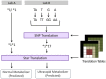
Existing approaches to managing genetic and genomic test results from external laboratories typically include filing of text reports within the electronic health record, making them unavailable in many cases for clinical decision support. Even when structured computable results are available, the lack of adopted standards requires considerations for processing the results into actionable knowledge, in addition to storage and management of the data. Here, we describe the design and implementation of an ancillary genomics system used to receive and process heterogeneous results from external laboratories, which returns a descriptive phenotype to the electronic health record in support of pharmacogenetic clinical decision support.
Keywords: electronic health record; genetic testing; pharmacogenomics.
© The Author(s) 2019. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Beyan T, Aydin Son Y.. Incorporation of personal single nucleotide polymorphism (SNP) data into a national level electronic health record for disease risk assessment, part 2: the incorporation of SNP into the national health information system of Turkey. JMIR Med Inform 2014; 2 (2): e17.. - PMC - PubMed
-
- Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM.. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin Pharmacol Ther 2017; 102 (2): 254–64. - PubMed

