Occurrence, function, and biosynthesis of mycofactocin
- PMID: 30778644
- PMCID: PMC6449177
- DOI: 10.1007/s00253-019-09684-4
Occurrence, function, and biosynthesis of mycofactocin
Abstract
Mycofactocin is a member of the rapidly growing class of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products. Although the mycofactocin biosynthetic pathway is widely distributed among Mycobacterial species, the structure, function, and biosynthesis of the pathway product remain unknown. This mini-review will discuss the current state of knowledge regarding the mycofactocin biosynthetic pathway. In particular, we focus on the architecture and distribution of the mycofactocin biosynthetic cluster, mftABCDEF, among the Actinobacteria phylum. We discuss the potential molecular and physiological role of mycofactocin. We review known biosynthetic steps involving MftA, MftB, MftC, and MftE and relate them to pyrroloquinoline quinone biosynthesis. Lastly, we propose the function of the remaining putative biosynthetic enzymes, MftD and MftF.
Keywords: Biosynthesis; Mycofactocin; Peptide modification; Redox cofactor; RiPP.
Conflict of interest statement
Conflicts of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Figures
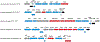


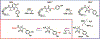
References
-
- Anthony C (2001) Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxidants Redox Signal 3:757–774 - PubMed
-
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160 . doi: 10.1039/C2NP20085F - DOI - PMC - PubMed
-
- Ayikpoe R, Ngendahimana T, Langton M, Bonitatibus S, Walker LM, Eaton SS, Eaton GR, Pandelia ME, Elliott SJ, Latham JA (2019) Spectroscopic and electrochemical characterization of the mycofactocin biosynthetic protein, MftC, provides insight into its redox flipping mechanism. Biohcemistry 10.1021/acs.biochem.8b01082 - DOI - PMC - PubMed
-
- Ayikpoe R, Salazar J, Majestic B, Latham JA (2018) Mycofactocin biosynthesis proceeds through 3-amino-5-[(p-hydroxyphenyl)methyl]-4,4-dimethyl-2-pyrrolidinone (AHDP); direct observation of MftE specificity toward MftA*. Biochemistry 57:5379–5383 . doi: 10.1021/acs.biochem.8b00816 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

