Predicting Postoperative Liver Dysfunction Based on Blood-Derived MicroRNA Signatures
- PMID: 30779441
- PMCID: PMC6593830
- DOI: 10.1002/hep.30572
Predicting Postoperative Liver Dysfunction Based on Blood-Derived MicroRNA Signatures
Abstract
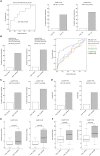
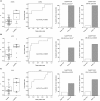
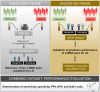
There is an urgent need for an easily assessable preoperative test to predict postoperative liver function recovery and thereby determine the optimal time point of liver resection, specifically as current markers are often expensive, time consuming, and invasive. Emerging evidence suggests that microRNA (miRNA) signatures represent potent diagnostic, prognostic, and treatment-response biomarkers for several diseases. Using next-generation sequencing as an unbiased systematic approach, 554 miRNAs were detected in preoperative plasma of 21 patients suffering from postoperative liver dysfunction (LD) after liver resection and 27 matched controls. Subsequently, we identified a miRNA signature-consisting of miRNAs 151a-5p, 192-5p, and 122-5p-that highly correlated with patients developing postoperative LD after liver resection. The predictive potential for postoperative LD was subsequently confirmed using real-time PCR in an independent validation cohort of 98 patients. Ultimately, a regression model of the two miRNA ratios 151a-5p to 192-5p and 122-5p to 151a-5p was found to reliably predict postoperative LD, severe morbidity, prolonged intensive care unit and hospital stays, and even mortality before an operation with a remarkable accuracy, thereby outperforming established markers of postoperative LD. Ultimately, we documented that miRNA ratios closely followed liver function recovery after partial hepatectomy. Conclusion: Our data demonstrate the clinical utility of an miRNA-based biomarker to support the selection of patients undergoing partial hepatectomy. The dynamical changes during liver function recovery indicate a possible role in individualized patient treatment. Thereby, our data might help to tailor surgical strategies to the specific risk profile of patients.
© 2019 The Authors. Hepatology published by Wiley Periodicals, Inc., on behalf of American Association for the Study of Liver Diseases.
Figures



Comment in
-
Reply.Hepatology. 2019 Sep;70(3):1082-1083. doi: 10.1002/hep.30682. Hepatology. 2019. PMID: 31032980 Free PMC article. No abstract available.
-
Letter to the Editor: Predicting Postoperative Liver Dysfunction Based on Blood Derived MicroRNA Signatures.Hepatology. 2019 Sep;70(3):1080-1081. doi: 10.1002/hep.30689. Hepatology. 2019. PMID: 31034633 No abstract available.
-
Letter to the Editor: Predicting Postoperative Liver Dysfunction Based on Blood-Derived MicroRNA Signatures.Hepatology. 2019 Sep;70(3):1081. doi: 10.1002/hep.30688. Hepatology. 2019. PMID: 31034655 No abstract available.
-
Letter to Editor: Association Between Postoperative Liver Dysfunction and Blood-Derived MicroRNA Signatures.Hepatology. 2019 Sep;70(3):1085-1086. doi: 10.1002/hep.30759. Hepatology. 2019. PMID: 31081219 No abstract available.
-
Reply.Hepatology. 2019 Sep;70(3):1086-1088. doi: 10.1002/hep.30764. Hepatology. 2019. PMID: 31081551 Free PMC article. No abstract available.
References
-
- Forbes SJ, Newsome PN. Liver regeneration—mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473‐485. - PubMed
-
- Lafaro K, Buettner S, Maqsood H, Wagner D, Bagante F, Spolverato G, et al. Defining post hepatectomy liver insufficiency: where do we stand? J Gastrointest Surg 2015;19:2079‐2092. - PubMed
-
- Qadan M, Garden OJ, Corvera CU, Visser BC. Management of postoperative hepatic failure. J Am Coll Surg 2016;222:195‐208. - PubMed
-
- Hackl M, Heilmeier U, Weilner S, Grillari J. Circulating microRNAs as novel biomarkers for bone diseases—complex signatures for multifactorial diseases? Mol Cell Endocrinol 2016;432:83‐95. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281‐297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

