Chromatin remodeler HELLS maintains glioma stem cells through E2F3 and MYC
- PMID: 30779712
- PMCID: PMC6483649
- DOI: 10.1172/jci.insight.126140
Chromatin remodeler HELLS maintains glioma stem cells through E2F3 and MYC
Abstract
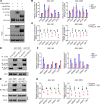
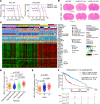
Glioblastomas, which contain stem cell-like glioblastoma stem cells (GSCs), are universally lethal cancers. While neural stem cells (NSCs) are usually quiescent, single-cell studies suggest that proliferating glioblastoma cells reside in the GSC population. Interrogating in silico glioma databases for epigenetic regulators that correlate with cell cycle regulation, we identified the chromatin remodeler HELLS as a potential target in glioblastoma. GSCs preferentially expressed HELLS compared with their differentiated tumor progeny and nonmalignant brain cells. Targeting HELLS disrupted GSC proliferation, survival, and self-renewal with induction of replication stress and DNA damage. Investigating potential molecular mechanisms downstream of HELLS revealed that HELLS interacted with the core oncogenic transcription factors, E2F3 and MYC, to regulate gene expression critical to GSC proliferation and maintenance. Supporting the interaction, HELLS expression strongly correlated with targets of E2F3 and MYC transcriptional activity in glioblastoma patients. The potential clinical significance of HELLS was reinforced by improved survival of tumor-bearing mice upon targeting HELLS and poor prognosis of glioma patients with elevated HELLS expression. Collectively, targeting HELLS may permit the functional disruption of the relatively undruggable MYC and E2F3 transcription factors and serve as a novel therapeutic paradigm for glioblastoma.
Keywords: Brain cancer; Oncology; Stem cells.
Conflict of interest statement
Figures


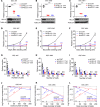
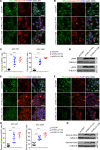
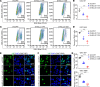
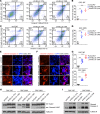

References
-
- Weller M, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS091080/NS/NINDS NIH HHS/United States
- R01 CA169117/CA/NCI NIH HHS/United States
- TL1 TR002549/TR/NCATS NIH HHS/United States
- F30 CA217065/CA/NCI NIH HHS/United States
- R01 NS099175/NS/NINDS NIH HHS/United States
- F30 CA217066/CA/NCI NIH HHS/United States
- R01 NS087913/NS/NINDS NIH HHS/United States
- R01 NS103434/NS/NINDS NIH HHS/United States
- R35 CA197718/CA/NCI NIH HHS/United States
- F30 CA203101/CA/NCI NIH HHS/United States
- R01 CA171652/CA/NCI NIH HHS/United States
- R01 NS089272/NS/NINDS NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- R01 CA154130/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

