Functional lability of RNA-dependent RNA polymerases in animals
- PMID: 30779744
- PMCID: PMC6396948
- DOI: 10.1371/journal.pgen.1007915
Functional lability of RNA-dependent RNA polymerases in animals
Abstract
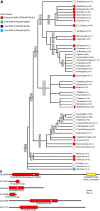
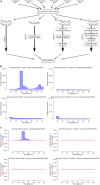
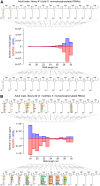
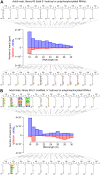
RNA interference (RNAi) requires RNA-dependent RNA polymerases (RdRPs) in many eukaryotes, and RNAi amplification constitutes the only known function for eukaryotic RdRPs. Yet in animals, classical model organisms can elicit RNAi without possessing RdRPs, and only nematode RNAi was shown to require RdRPs. Here we show that RdRP genes are much more common in animals than previously thought, even in insects, where they had been assumed not to exist. RdRP genes were present in the ancestors of numerous clades, and they were subsequently lost at a high frequency. In order to probe the function of RdRPs in a deuterostome (the cephalochordate Branchiostoma lanceolatum), we performed high-throughput analyses of small RNAs from various Branchiostoma developmental stages. Our results show that Branchiostoma RdRPs do not appear to participate in RNAi: we did not detect any candidate small RNA population exhibiting classical siRNA length or sequence features. Our results show that RdRPs have been independently lost in dozens of animal clades, and even in a clade where they have been conserved (cephalochordates) their function in RNAi amplification is not preserved. Such a dramatic functional variability reveals an unexpected plasticity in RNA silencing pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
RNA-dependent RNA polymerases in RNA silencing.Biol Chem. 2011 Apr;392(4):299-304. doi: 10.1515/BC.2011.035. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294682 Review.
-
Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups.Gene. 2009 Nov 1;447(1):29-39. doi: 10.1016/j.gene.2009.07.004. Epub 2009 Jul 17. Gene. 2009. PMID: 19616606
-
Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides.Mol Microbiol. 2012 Jan;83(2):379-94. doi: 10.1111/j.1365-2958.2011.07939.x. Epub 2011 Dec 19. Mol Microbiol. 2012. PMID: 22141923
-
RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase.RNA. 2003 Feb;9(2):187-92. doi: 10.1261/rna.2860603. RNA. 2003. PMID: 12554861 Free PMC article.
-
RNA-dependent RNA polymerases, viruses, and RNA silencing.Science. 2002 May 17;296(5571):1270-3. doi: 10.1126/science.1069132. Science. 2002. PMID: 12016304 Review.
Cited by
-
RNAi-Based Functional Genomics in Hemiptera.Insects. 2020 Aug 20;11(9):557. doi: 10.3390/insects11090557. Insects. 2020. PMID: 32825516 Free PMC article. Review.
-
Insights Into an Unexplored Component of the Mosquito Repeatome: Distribution and Variability of Viral Sequences Integrated Into the Genome of the Arboviral Vector Aedes albopictus.Front Genet. 2019 Feb 12;10:93. doi: 10.3389/fgene.2019.00093. eCollection 2019. Front Genet. 2019. PMID: 30809249 Free PMC article.
-
Target-specific requirements for RNA interference can arise through restricted RNA amplification despite the lack of specialized pathways.Elife. 2024 Aug 20;13:RP97487. doi: 10.7554/eLife.97487. Elife. 2024. PMID: 39161220 Free PMC article.
-
Toward a Categorization of Virus-ncRNA Interactions in the World of RNA to Disentangle the Tiny Secrets of Dengue Virus.Viruses. 2024 May 18;16(5):804. doi: 10.3390/v16050804. Viruses. 2024. PMID: 38793685 Free PMC article. Review.
-
A novel eukaryotic RdRP-dependent small RNA pathway represses antiviral immunity by controlling an ERK pathway component in the black-legged tick.PLoS One. 2023 Mar 30;18(3):e0281195. doi: 10.1371/journal.pone.0281195. eCollection 2023. PLoS One. 2023. PMID: 36996253 Free PMC article.
References
-
- Schiebel W, Haas B, Marinković S, Klanner A, Sänger HL. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J Biol Chem. 1993;268(16):11858–11867. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

