Identification of HIV gp41-specific antibodies that mediate killing of infected cells
- PMID: 30779811
- PMCID: PMC6396944
- DOI: 10.1371/journal.ppat.1007572
Identification of HIV gp41-specific antibodies that mediate killing of infected cells
Abstract
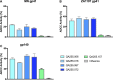
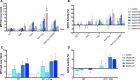
Antibodies that mediate killing of HIV-infected cells through antibody-dependent cellular cytotoxicity (ADCC) have been implicated in protection from HIV infection and disease progression. Despite these observations, these types of HIV antibodies are understudied compared to neutralizing antibodies. Here we describe four monoclonal antibodies (mAbs) obtained from one individual that target the HIV transmembrane protein, gp41, and mediate ADCC activity. These four mAbs arose from independent B cell lineages suggesting that in this individual, multiple B cell responses were induced by the gp41 antigen. Competition and phage peptide display mapping experiments suggested that two of the mAbs target epitopes in the cysteine loop that are highly conserved and a common target of HIV gp41-specific antibodies. The amino acid sequences that bind these mAbs are overlapping but distinct. The two other mAbs were competed by mAbs that target the C-terminal heptad repeat (CHR) and the fusion peptide proximal region (FPPR) and appear to both target a similar unique conformational epitope. These gp41-specific mAbs mediated killing of infected cells that express high levels of Env due to either pre-treatment with interferon or deletion of vpu to increase levels of BST-2/Tetherin. They also mediate killing of target cells coated with various forms of the gp41 protein, including full-length gp41, gp41 ectodomain or a mimetic of the gp41 stump. Unlike many ADCC mAbs that target HIV gp120, these gp41-mAbs are not dependent on Env structural changes associated with membrane-bound CD4 interaction. Overall, the characterization of these four new mAbs that target gp41 and mediate ADCC provides evidence for diverse gp41 B cell lineages with overlapping but distinct epitopes within an individual. Such antibodies that can target various forms of envelope protein could represent a common response to a relatively conserved HIV epitope for a vaccine.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

