Blood-Derived DNA Methylation Signatures of Crohn's Disease and Severity of Intestinal Inflammation
- PMID: 30779925
- PMCID: PMC6529254
- DOI: 10.1053/j.gastro.2019.01.270
Blood-Derived DNA Methylation Signatures of Crohn's Disease and Severity of Intestinal Inflammation
Abstract
Background & aims: Crohn's disease is a relapsing and remitting inflammatory disorder with a variable clinical course. Although most patients present with an inflammatory phenotype (B1), approximately 20% of patients rapidly progress to complicated disease, which includes stricturing (B2), within 5 years. We analyzed DNA methylation patterns in blood samples of pediatric patients with Crohn's disease at diagnosis and later time points to identify changes that associate with and might contribute to disease development and progression.
Methods: We obtained blood samples from 164 pediatric patients (1-17 years old) with Crohn's disease (B1 or B2) who participated in a North American study and were followed for 5 years. Participants without intestinal inflammation or symptoms served as controls (n = 74). DNA methylation patterns were analyzed in samples collected at time of diagnosis and 1-3 years later at approximately 850,000 sites. We used genetic association and the concept of Mendelian randomization to identify changes in DNA methylation patterns that might contribute to the development of or result from Crohn's disease.
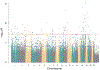
Results: We identified 1189 5'-cytosine-phosphate-guanosine-3' (CpG) sites that were differentially methylated between patients with Crohn's disease (at diagnosis) and controls. Methylation changes at these sites correlated with plasma levels of C-reactive protein. A comparison of methylation profiles of DNA collected at diagnosis of Crohn's disease vs during the follow-up period showed that, during treatment, alterations identified in methylation profiles at the time of diagnosis of Crohn's disease more closely resembled patterns observed in controls, irrespective of disease progression to B2. We identified methylation changes at 3 CpG sites that might contribute to the development of Crohn's disease. Most CpG methylation changes associated with Crohn's disease disappeared with treatment of inflammation and might be a result of Crohn's disease.
Conclusions: Methylation patterns observed in blood samples from patients with Crohn's disease accompany acute inflammation; with treatment, these change to resemble methylation patterns observed in patients without intestinal inflammation. These findings indicate that Crohn's disease-associated patterns of DNA methylation observed in blood samples are a result of the inflammatory features of the disease and are less likely to contribute to disease development or progression.
Keywords: Children; Epigenetic Alteration; Inflammatory Bowel Disease; Risk Stratification and Identification of Immunogenetic and Microbial Markers of Rapid Disease Progression in Children with Crohn's Disease (RISK) Study.
Copyright © 2019 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest
The authors disclose no conflicts.
Figures

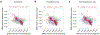


Comment in
-
The value of blood derived DNA methylation signatures in advancing our understanding of Crohn's Disease pathogenesis.Transl Gastroenterol Hepatol. 2019 Aug 20;4:60. doi: 10.21037/tgh.2019.08.07. eCollection 2019. Transl Gastroenterol Hepatol. 2019. PMID: 31559341 Free PMC article. No abstract available.
-
Blood-based DNA methylation in Crohn's disease and severity of intestinal inflammation.Transl Gastroenterol Hepatol. 2019 Nov 26;4:76. doi: 10.21037/tgh.2019.10.03. eCollection 2019. Transl Gastroenterol Hepatol. 2019. PMID: 31872140 Free PMC article. No abstract available.
References
-
- Van Limbergen J, Russell RK, Drummond HE, et al. Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology 2008; 135:1114–22. - PubMed
-
- Ruel J, Ruane D, Mehandru S, et al. IBD across the age spectrum: is it the same disease? Nat Rev Gastroenterol Hepatol 2014;11:88–98. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

