Trends in the Design of New Isobaric Labeling Reagents for Quantitative Proteomics
- PMID: 30781343
- PMCID: PMC6412310
- DOI: 10.3390/molecules24040701
Trends in the Design of New Isobaric Labeling Reagents for Quantitative Proteomics
Abstract
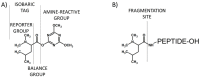
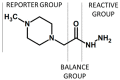
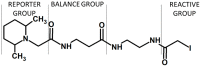
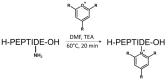
Modern mass spectrometry is one of the most frequently used methods of quantitative proteomics, enabling determination of the amount of peptides in a sample. Although mass spectrometry is not inherently a quantitative method due to differences in the ionization efficiency of various analytes, the application of isotope-coded labeling allows relative quantification of proteins and proteins. Over the past decade, a new method for derivatization of tryptic peptides using isobaric labels has been proposed. The labels consist of reporter and balanced groups. They have the same molecular weights and chemical properties, but differ in the distribution of stable heavy isotopes. These tags are designed in such a way that during high energy collision induced dissociation (CID) by tandem mass spectrometry, the isobaric tag is fragmented in the specific linker region, yielding reporter ions with different masses. The mass shifts among the reporter groups are compensated by the balancing groups so that the overall mass is the same for all forms of the reagent. Samples of peptides are labeled with the isobaric mass tags in parallel and combined for analysis. Quantification of individual peptides is achieved by comparing the intensity of reporter ions in the tandem mass (MS/MS) spectra. Isobaric markers have found a wide range of potential applications in proteomics. However, the currently available isobaric labeling reagents have some drawbacks, such as high cost of production, insufficient selectivity of the derivatization, and relatively limited enhancement of sensitivity of the analysis. Therefore, efforts have been devoted to the development of new isobaric markers with increased usability. The search for new isobaric markers is focused on developing a more selective method of introducing a tag into a peptide molecule, increasing the multiplexicity of markers, lowering the cost of synthesis, and increasing the sensitivity of measurement by using ionization tags containing quaternary ammonium salts. Here, the trends in the design of new isobaric labeling reagents for quantitative proteomics isobaric derivatization strategies in proteomics are reviewed, with a particular emphasis on isobaric ionization tags. The presented review focused on different types of isobaric reagents used in quantitative proteomics, their chemistry, and advantages offer by their application.
Keywords: ESI-MS; ionization enhancers; isobaric labeling reagents; quantitative proteomics; quaternary ammonium salts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
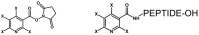
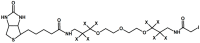
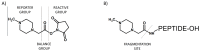
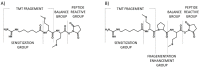
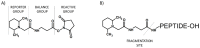
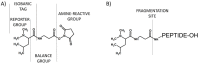
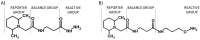
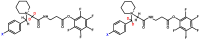
Similar articles
-
Quaternary ammonium isobaric tag for a relative and absolute quantification of peptides.J Mass Spectrom. 2018 Feb;53(2):115-123. doi: 10.1002/jms.4040. J Mass Spectrom. 2018. PMID: 29087004
-
Reagents for Isobaric Labeling Peptides in Quantitative Proteomics.Anal Chem. 2018 Nov 6;90(21):12366-12371. doi: 10.1021/acs.analchem.8b00321. Epub 2018 Oct 16. Anal Chem. 2018. PMID: 30260629
-
Quantitative proteome analysis using isobaric peptide termini labeling (IPTL).Methods Mol Biol. 2011;753:65-76. doi: 10.1007/978-1-61779-148-2_5. Methods Mol Biol. 2011. PMID: 21604116
-
Standardization approaches in absolute quantitative proteomics with mass spectrometry.Mass Spectrom Rev. 2018 Nov;37(6):715-737. doi: 10.1002/mas.21542. Epub 2017 Jul 31. Mass Spectrom Rev. 2018. PMID: 28758227 Review.
-
Multi-functional MBIT for peptide tandem mass spectrometry.Mass Spectrom Rev. 2015 Mar-Apr;34(2):209-18. doi: 10.1002/mas.21435. Epub 2014 May 28. Mass Spectrom Rev. 2015. PMID: 24872020 Review.
Cited by
-
Proteomics: A Tool to Study Platelet Function.Int J Mol Sci. 2021 Apr 30;22(9):4776. doi: 10.3390/ijms22094776. Int J Mol Sci. 2021. PMID: 33946341 Free PMC article. Review.
-
Proteomics and Bioinformatics Analysis of Cartilage in Post-Traumatic Osteoarthritis in a Mini-Pig Model of Anterior Cruciate Ligament Repair.Med Sci Monit. 2020 Jan 9;26:e920104. doi: 10.12659/MSM.920104. Med Sci Monit. 2020. PMID: 31916546 Free PMC article.
-
Statistical Detection of Differentially Abundant Proteins in Experiments with Repeated Measures Designs and Isobaric Labeling.J Proteome Res. 2023 Aug 4;22(8):2641-2659. doi: 10.1021/acs.jproteome.3c00155. Epub 2023 Jul 19. J Proteome Res. 2023. PMID: 37467362 Free PMC article.
-
Enrichment of Cysteine-Containing Peptide by On-Resin Capturing and Fixed Charge Tag Derivatization for Sensitive ESI-MS Detection.Molecules. 2020 Mar 18;25(6):1372. doi: 10.3390/molecules25061372. Molecules. 2020. PMID: 32197294 Free PMC article.
-
Detection of Podocin in Human Urine Sediment Samples by Charge Derivatization and LC-MS-MRM Method.Int J Mol Sci. 2020 May 2;21(9):3225. doi: 10.3390/ijms21093225. Int J Mol Sci. 2020. PMID: 32370166 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

