PheWAS-Based Systems Genetics Methods for Anti-Breast Cancer Drug Discovery
- PMID: 30781719
- PMCID: PMC6409623
- DOI: 10.3390/genes10020154
PheWAS-Based Systems Genetics Methods for Anti-Breast Cancer Drug Discovery
Abstract
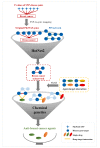
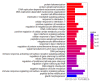
Breast cancer is a high-risk disease worldwide. For such complex diseases that are induced by multiple pathogenic genes, determining how to establish an effective drug discovery strategy is a challenge. In recent years, a large amount of genetic data has accumulated, particularly in the genome-wide identification of disorder genes. However, understanding how to use these data efficiently for pathogenesis elucidation and drug discovery is still a problem because the gene⁻disease links that are identified by high-throughput techniques such as phenome-wide association studies (PheWASs) are usually too weak to have biological significance. Systems genetics is a thriving area of study that aims to understand genetic interactions on a genome-wide scale. In this study, we aimed to establish two effective strategies for identifying breast cancer genes based on the systems genetics algorithm. As a result, we found that the GeneRank-based strategy, which combines the prognostic phenotype-based gene-dependent network with the phenotypic-related PheWAS data, can promote the identification of breast cancer genes and the discovery of anti-breast cancer drugs.
Keywords: PheWAS; breast cancer; drug discovery; systems genetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Genome-wide pathogenesis interpretation using a heat diffusion-based systems genetics method and implications for gene function annotation.Mol Genet Genomic Med. 2020 Oct;8(10):e1456. doi: 10.1002/mgg3.1456. Epub 2020 Sep 1. Mol Genet Genomic Med. 2020. PMID: 32869547 Free PMC article.
-
Pharmacogenomic Discovery to Function and Mechanism: Breast Cancer as a Case Study.Clin Pharmacol Ther. 2018 Feb;103(2):243-252. doi: 10.1002/cpt.915. Epub 2017 Dec 1. Clin Pharmacol Ther. 2018. PMID: 29052219 Free PMC article.
-
Genetics and genome-wide association studies: surgery-guided algorithm and promise for future breast cancer personalized surgery.Expert Rev Mol Diagn. 2008 Sep;8(5):587-97. doi: 10.1586/14737159.8.5.587. Expert Rev Mol Diagn. 2008. PMID: 18785807
-
Nanobiotechnological Approaches to Overcome Drug Resistance in Breast Cancer.Curr Cancer Drug Targets. 2015;15(7):544-62. doi: 10.2174/1568009615666150706102842. Curr Cancer Drug Targets. 2015. PMID: 26143946 Review.
-
Patient-derived tumour xenografts for breast cancer drug discovery.Endocr Relat Cancer. 2016 Dec;23(12):T259-T270. doi: 10.1530/ERC-16-0251. Epub 2016 Oct 4. Endocr Relat Cancer. 2016. PMID: 27702751 Free PMC article. Review.
Cited by
-
Unsupervised deep learning of electrocardiograms enables scalable human disease profiling.NPJ Digit Med. 2025 Jan 12;8(1):23. doi: 10.1038/s41746-024-01418-9. NPJ Digit Med. 2025. PMID: 39799251 Free PMC article.
-
LINA: A Linearizing Neural Network Architecture for Accurate First-Order and Second-Order Interpretations.IEEE Access. 2022;10:36166-36176. doi: 10.1109/access.2022.3163257. Epub 2022 Mar 30. IEEE Access. 2022. PMID: 35462722 Free PMC article.
References
-
- Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., Schacht A.L. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

