The Modern View of B Chromosomes Under the Impact of High Scale Omics Analyses
- PMID: 30781835
- PMCID: PMC6406668
- DOI: 10.3390/cells8020156
The Modern View of B Chromosomes Under the Impact of High Scale Omics Analyses
Abstract
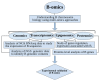
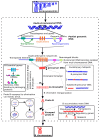
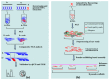
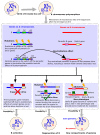
Supernumerary B chromosomes (Bs) are extra karyotype units in addition to A chromosomes, and are found in some fungi and thousands of animals and plant species. Bs are uniquely characterized due to their non-Mendelian inheritance, and represent one of the best examples of genomic conflict. Over the last decades, their genetic composition, function and evolution have remained an unresolved query, although a few successful attempts have been made to address these phenomena. A classical concept based on cytogenetics and genetics is that Bs are selfish and abundant with DNA repeats and transposons, and in most cases, they do not carry any function. However, recently, the modern quantum development of high scale multi-omics techniques has shifted B research towards a new-born field that we call "B-omics". We review the recent literature and add novel perspectives to the B research, discussing the role of new technologies to understand the mechanistic perspectives of the molecular evolution and function of Bs. The modern view states that B chromosomes are enriched with genes for many significant biological functions, including but not limited to the interesting set of genes related to cell cycle and chromosome structure. Furthermore, the presence of B chromosomes could favor genomic rearrangements and influence the nuclear environment affecting the function of other chromatin regions. We hypothesize that B chromosomes might play a key function in driving their transmission and maintenance inside the cell, as well as offer an extra genomic compartment for evolution.
Keywords: cytogenetics; evolution; genes; genome; next generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Longley A.E. Supernumerary chromosomes in Zea mays. J. Agric. Res. 1927;35:769–784.
-
- Randolph L.F. Types of supernumerary chromosomes in maize. Anat. Rec. 1928;41:102.
-
- Wilson E.B. The supernumerary chromosomes of Hemiptera. Sci. New Ser. 1907
-
- Stevens N.M. The chromosomes in Diabrotica vittata, Diabrotica soror and Diabrotica punctata. A contribution to the literature on heterochromosomes and sex determination. J. Exp. Zool. 1908;5:453–470. doi: 10.1002/jez.1400050402. - DOI
-
- Beukeboom L.W. Bewildering Bs: An impression of the 1st B-Chromosome Conference. Heredity. 1994;73:328–336. doi: 10.1038/hdy.1994.140. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

