Genomic evidence for shared common ancestry of East African hunting-gathering populations and insights into local adaptation
- PMID: 30782801
- PMCID: PMC6410815
- DOI: 10.1073/pnas.1817678116
Genomic evidence for shared common ancestry of East African hunting-gathering populations and insights into local adaptation
Abstract
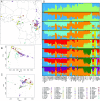
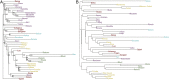
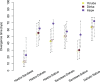
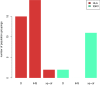
Anatomically modern humans arose in Africa ∼300,000 years ago, but the demographic and adaptive histories of African populations are not well-characterized. Here, we have generated a genome-wide dataset from 840 Africans, residing in western, eastern, southern, and northern Africa, belonging to 50 ethnicities, and speaking languages belonging to four language families. In addition to agriculturalists and pastoralists, our study includes 16 populations that practice, or until recently have practiced, a hunting-gathering (HG) lifestyle. We observe that genetic structure in Africa is broadly correlated not only with geography, but to a lesser extent, with linguistic affiliation and subsistence strategy. Four East African HG (EHG) populations that are geographically distant from each other show evidence of common ancestry: the Hadza and Sandawe in Tanzania, who speak languages with clicks classified as Khoisan; the Dahalo in Kenya, whose language has remnant clicks; and the Sabue in Ethiopia, who speak an unclassified language. Additionally, we observed common ancestry between central African rainforest HGs and southern African San, the latter of whom speak languages with clicks classified as Khoisan. With the exception of the EHG, central African rainforest HGs, and San, other HG groups in Africa appear genetically similar to neighboring agriculturalist or pastoralist populations. We additionally demonstrate that infectious disease, immune response, and diet have played important roles in the adaptive landscape of African history. However, while the broad biological processes involved in recent human adaptation in Africa are often consistent across populations, the specific loci affected by selective pressures more often vary across populations.
Keywords: African diversity; African hunter-gatherers; human evolution; natural selection; population genetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. - PubMed
-
- McDermott F, et al. New Late-Pleistocene uranium–thorium and ESR dates for the Singa hominid (Sudan) J Hum Evol. 1996;31:507–516.
-
- Hublin JJ, et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature. 2017;546:289–292. - PubMed
-
- Mcbrearty S, Brooks AS. The revolution that wasn’t: A new interpretation of the origin of modern human behavior. J Hum Evol. 2000;39:453–563. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

