Processing generates 3' ends of RNA masking transcription termination events in prokaryotes
- PMID: 30782818
- PMCID: PMC6410799
- DOI: 10.1073/pnas.1813181116
Processing generates 3' ends of RNA masking transcription termination events in prokaryotes
Abstract
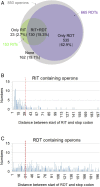
Two kinds of signal-dependent transcription termination and RNA release mechanisms have been established in prokaryotes in vitro by: (i) binding of Rho to cytidine-rich nascent RNA [Rho-dependent termination (RDT)], and (ii) the formation of a hairpin structure in the nascent RNA, ending predominantly with uridine residues [Rho-independent termination (RIT)]. As shown here, the two signals act independently of each other and can be regulated (suppressed) by translation-transcription coupling in vivo. When not suppressed, both RIT- and RDT-mediated transcription termination do occur, but ribonucleolytic processing generates defined new 3' ends in the terminated RNA molecules. The actual termination events at the end of transcription units are masked by generation of new processed 3' RNA ends; thus the in vivo 3' ends do not define termination sites. We predict generation of 3' ends of mRNA by processing is a common phenomenon in prokaryotes as is the case in eukaryotes.
Keywords: RNA processing; gal operon; transcription termination; translation–transcription coupling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

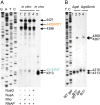
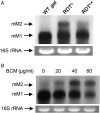
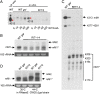
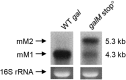
References
-
- Richardson JP. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991;64:1047–1049. - PubMed
-
- Adhya S, Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. - PubMed
-
- Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. - PubMed
-
- Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–952. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

