Thermodynamic insights into an interaction between ACYL-CoA-BINDING PROTEIN2 and LYSOPHOSPHOLIPASE2 in Arabidopsis
- PMID: 30782848
- PMCID: PMC6484133
- DOI: 10.1074/jbc.RA118.006876
Thermodynamic insights into an interaction between ACYL-CoA-BINDING PROTEIN2 and LYSOPHOSPHOLIPASE2 in Arabidopsis
Abstract
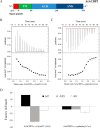
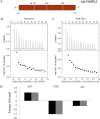
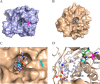
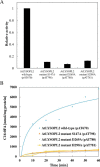
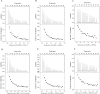
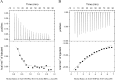
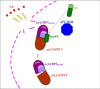
Lysophospholipids (LPLs) are important lipid-signaling molecules in plants, of which lysophosphatidylcholine (lysoPC) is one of the most well-characterized LPLs, having important roles in plant stress responses. It is broken down by lysophospholipases, but the molecular mechanism involved in lysoPC degradation is unclear. Recombinant Arabidopsis thaliana ACYL-CoA-BINDING PROTEIN2 (AtACBP2) has been reported to bind lysoPC via its acyl-CoA-binding domain and also LYSOPHOSPHOLIPASE 2 (AtLYSOPL2) via its ankyrin repeats in vitro To investigate the interactions of AtACBP2 with AtLYSOPL2 and lysoPC in more detail, we conducted isothermal titration calorimetry with AtACBP270-354, an AtACBP2 derivative consisting of amino acids 70-354, containing both the acyl-CoA-binding domain and ankyrin repeats. We observed that the interactions of AtACBP270-354 with AtLYSOPL2 and lysoPC were both endothermic, favored by solvation entropy and opposed by enthalpy, with dissociation constants in the micromolar range. Of note, three AtLYSOPL2 catalytic triad mutant proteins (S147A, D268A, and H298A) bound lysoPC only weakly, with an exothermic burst and dissociation constants in the millimolar range. Furthermore, the binding affinity of lysoPC-premixed AtACBP270-354 to AtLYSOPL2 was 10-fold higher than that of AtACBP270-354 alone to AtLYSOPL2. We conclude that AtACBP2 may play a role in facilitating a direct interaction between AtLYSOPL2 and lysoPC. Our results suggest that AtACBP270-354 probably binds to lysoPC through a hydrophobic interface that enhances a hydrotropic interaction of AtACBP270-354 with AtLYSOPL2 and thereby facilitates AtLYSOPL2's lysophospholipase function.
Keywords: ankyrin; enzyme mutation; isothermal titration calorimetry (ITC); lipid metabolism; lysophosphatidylcholine; lysophospholipid; molecular docking; protein–protein interaction; structural model; thermodynamics.
© 2019 Miao et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
References
-
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H. E., Rajashekar C. B., Williams T. D., and Wang X. (2002) Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002 10.1074/jbc.M205375200 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

