The Possibility of Systematic Research Fraud Targeting Under-Studied Human Genes: Causes, Consequences, and Potential Solutions
- PMID: 30783377
- PMCID: PMC6366001
- DOI: 10.1177/1177271919829162
The Possibility of Systematic Research Fraud Targeting Under-Studied Human Genes: Causes, Consequences, and Potential Solutions
Abstract
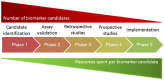
A major reason for biomarker failure is the selection of candidate biomarkers based on inaccurate or incorrect published results. Incorrect research results leading to the selection of unproductive biomarker candidates are largely considered to stem from unintentional research errors. The additional possibility that biomarker research may be actively misdirected by research fraud has been given comparatively little consideration. This review discusses what we believe to be a new threat to biomarker research, namely, the possible systematic production of fraudulent gene knockdown studies that target under-studied human genes. We describe how fraudulent papers may be produced in series by paper mills using what we have described as a 'theme and variations' model, which could also be considered a form of salami slicing. We describe features of these single-gene knockdown publications that may allow them to evade detection by journal editors, peer reviewers, and readers. We then propose a number of approaches to facilitate their detection, including improved awareness of the features of publications constructed in series, broader requirements to post submitted manuscripts to preprint servers, and the use of semi-automated literature screening tools. These approaches may collectively improve the detection of fraudulent studies that might otherwise impede future biomarker research.
Keywords: Biomarkers; cancer; gene knockdown techniques; paper mill; research fraud; salami publication; under-studied gene.
Conflict of interest statement
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



References
-
- de Gramont A, Watson S, Ellis LM, et al. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol. 2015;12:197–212. - PubMed
-
- Duffy MJ, Sturgeon CM, Sölétormos G, et al. Validation of new cancer biomarkers: a position statement from the European group on tumor markers. Clin Chem. 2015;61:809–820. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

