Embracing heterogeneity: coalescing the Tree of Life and the future of phylogenomics
- PMID: 30783571
- PMCID: PMC6378093
- DOI: 10.7717/peerj.6399
Embracing heterogeneity: coalescing the Tree of Life and the future of phylogenomics
Abstract
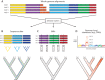
Building the Tree of Life (ToL) is a major challenge of modern biology, requiring advances in cyberinfrastructure, data collection, theory, and more. Here, we argue that phylogenomics stands to benefit by embracing the many heterogeneous genomic signals emerging from the first decade of large-scale phylogenetic analysis spawned by high-throughput sequencing (HTS). Such signals include those most commonly encountered in phylogenomic datasets, such as incomplete lineage sorting, but also those reticulate processes emerging with greater frequency, such as recombination and introgression. Here we focus specifically on how phylogenetic methods can accommodate the heterogeneity incurred by such population genetic processes; we do not discuss phylogenetic methods that ignore such processes, such as concatenation or supermatrix approaches or supertrees. We suggest that methods of data acquisition and the types of markers used in phylogenomics will remain restricted until a posteriori methods of marker choice are made possible with routine whole-genome sequencing of taxa of interest. We discuss limitations and potential extensions of a model supporting innovation in phylogenomics today, the multispecies coalescent model (MSC). Macroevolutionary models that use phylogenies, such as character mapping, often ignore the heterogeneity on which building phylogenies increasingly rely and suggest that assimilating such heterogeneity is an important goal moving forward. Finally, we argue that an integrative cyberinfrastructure linking all steps of the process of building the ToL, from specimen acquisition in the field to publication and tracking of phylogenomic data, as well as a culture that values contributors at each step, are essential for progress.
Keywords: Gene flow; Genome; Multispecies coalescent model; Retroelement; Speciation; Transcriptome.
Conflict of interest statement
Alexander Schliep and Scott V. Edwards are Academic Editors for PeerJ.
Figures
References
-
- Adamczak R, Miloś P. U-statistics of Ornstein-Uhlenbeck branching particle system. Journal of Theoretical Probability. 2014;27(4):1071–1111. doi: 10.1007/s10959-013-0503-2. - DOI
-
- Adamczak R, Miloś P. CLT for Ornstein-Uhlenbeck branching particle system. Electronic Journal of Probability. 2015;20:1–35. doi: 10.1214/EJP.v20-4233. - DOI
-
- Ané C. Analysis of comparative data with hierarchical autocorrelation. Annals of Applied Statistics. 2008;2(3):1078–1102. doi: 10.1214/08-AOAS173. - DOI
LinkOut - more resources
Full Text Sources

