MRE11 UFMylation promotes ATM activation
- PMID: 30783677
- PMCID: PMC6486557
- DOI: 10.1093/nar/gkz110
MRE11 UFMylation promotes ATM activation
Erratum in
-
Correction to 'MRE11 UFMylation promotes ATM activation'.Nucleic Acids Res. 2024 Oct 14;52(18):11412. doi: 10.1093/nar/gkae802. Nucleic Acids Res. 2024. PMID: 39268582 Free PMC article. No abstract available.
Abstract
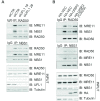
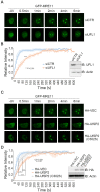
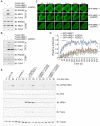
A proper DNA damage response (DDR) is essential to maintain genome integrity and prevent tumorigenesis. DNA double-strand breaks (DSBs) are the most toxic DNA lesion and their repair is orchestrated by the ATM kinase. ATM is activated via the MRE11-RAD50-NBS1 (MRN) complex along with its autophosphorylation at S1981 and acetylation at K3106. Activated ATM rapidly phosphorylates a vast number of substrates in local chromatin, providing a scaffold for the assembly of higher-order complexes that can repair damaged DNA. While reversible ubiquitination has an important role in the DSB response, modification of the newly identified ubiquitin-like protein ubiquitin-fold modifier 1 and the function of UFMylation in the DDR is largely unknown. Here, we found that MRE11 is UFMylated on K282 and this UFMylation is required for the MRN complex formation under unperturbed conditions and DSB-induced optimal ATM activation, homologous recombination-mediated repair and genome integrity. A pathogenic mutation MRE11(G285C) identified in uterine endometrioid carcinoma exhibited a similar cellular phenotype as the UFMylation-defective mutant MRE11(K282R). Taken together, MRE11 UFMylation promotes ATM activation, DSB repair and genome stability, and potentially serves as a therapeutic target.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Lee J.H., Paull T.T.. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004; 304:93–96. - PubMed
-
- Lee J.H., Paull T.T.. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005; 308:551–554. - PubMed
-
- Lee J.H., Paull T.T.. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007; 26:7741–7748. - PubMed
-
- Bakkenist C.J., Kastan M.B.. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003; 421:499–506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

