Myosin IIA drives membrane bleb retraction
- PMID: 30785846
- PMCID: PMC6724514
- DOI: 10.1091/mbc.E18-11-0752
Myosin IIA drives membrane bleb retraction
Abstract
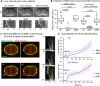
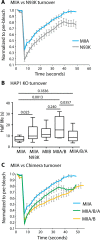
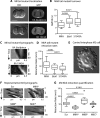
Membrane blebs are specialized cellular protrusions that play diverse roles in processes such as cell division and cell migration. Blebbing can be divided into three distinct phases: bleb nucleation, bleb growth, and bleb retraction. Following nucleation and bleb growth, the actin cortex, comprising actin, cross-linking proteins, and nonmuscle myosin II (MII), begins to reassemble on the membrane. MII then drives the final phase, bleb retraction, which results in reintegration of the bleb into the cellular cortex. There are three MII paralogues with distinct biophysical properties expressed in mammalian cells: MIIA, MIIB, and MIIC. Here we show that MIIA specifically drives bleb retraction during cytokinesis. The motor domain and regulation of the nonhelical tailpiece of MIIA both contribute to its ability to drive bleb retraction. These experiments have also revealed a relationship between faster turnover of MIIA at the cortex and its ability to drive bleb retraction.
Figures


References
-
- Blaser H, Reichman-Fried M, Castanon I, Dumstrei K, Marlow FL, Kawakami K, Solnica-Krezel L, Heisenberg C-P, Raz E. (2006). Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev Cell , 613–627. - PubMed
-
- Bluteau D, Glembotsky AC, Raimbault A, Balayn N, Gilles L, Rameau P, Nurden P, Alessi MC, Debili N, Vainchenker W, et al. (2012). Dysmegakaryopoiesis of FPD/AML pedigrees with constitutional RUNX1 mutations is linked to myosin II deregulated expression. Blood , 2708–2718. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

