Choice between 1- and 2-furrow cytokinesis in Caenorhabditis elegans embryos with tripolar spindles
- PMID: 30785847
- PMCID: PMC6727771
- DOI: 10.1091/mbc.E19-01-0075
Choice between 1- and 2-furrow cytokinesis in Caenorhabditis elegans embryos with tripolar spindles
Abstract
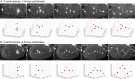
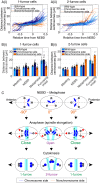
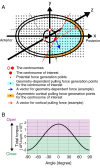
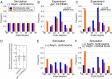
Excessive centrosomes often lead to multipolar spindles, and thus probably to multipolar mitosis and aneuploidy. In Caenorhabditis elegans, ∼70% of the paternal emb-27APC6 mutant embryonic cells contained more than two centrosomes and formed multipolar spindles. However, only ~30% of the cells with tripolar spindles formed two cytokinetic furrows. The rest formed one furrow, similar to normal cells. To investigate the mechanism via which cells avoid forming two cytokinetic furrows even with a tripolar spindle, we conducted live-cell imaging in emb-27APC6 mutant cells. We observed that the chromatids were aligned on only two of the three sides of the tripolar spindle, and the angle of the tripolar spindle relative to the long axis of the cell correlated with the number of cytokinetic furrows. Our numerical modeling showed that the combination of cell shape, cortical pulling forces, and heterogeneity of centrosome size determines whether cells with a tripolar spindle form one or two cytokinetic furrows.
Figures



References
-
- Boveri T. (2008). Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci (suppl 1), 1–84. - PubMed
-
- Bringmann H, Hyman AA. (2005). A cytokinesis furrow is positioned by two consecutive signals. Nature , 731–734. - PubMed
-
- Brinkley BR. (2001). Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol , 18–21. - PubMed
-
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gönczy P. (2003). Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science , 1957–1961. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

