Machine learning analysis of gene expression data reveals novel diagnostic and prognostic biomarkers and identifies therapeutic targets for soft tissue sarcomas
- PMID: 30785874
- PMCID: PMC6398862
- DOI: 10.1371/journal.pcbi.1006826
Machine learning analysis of gene expression data reveals novel diagnostic and prognostic biomarkers and identifies therapeutic targets for soft tissue sarcomas
Abstract
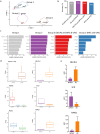
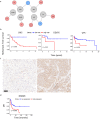
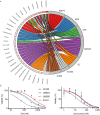
Based on morphology it is often challenging to distinguish between the many different soft tissue sarcoma subtypes. Moreover, outcome of disease is highly variable even between patients with the same disease. Machine learning on transcriptome sequencing data could be a valuable new tool to understand differences between and within entities. Here we used machine learning analysis to identify novel diagnostic and prognostic markers and therapeutic targets for soft tissue sarcomas. Gene expression data was used from the Cancer Genome Atlas, the Genotype-Tissue Expression project and the French Sarcoma Group. We identified three groups of tumors that overlap in their molecular profiles as seen with unsupervised t-Distributed Stochastic Neighbor Embedding clustering and a deep neural network. The three groups corresponded to subtypes that are morphologically overlapping. Using a random forest algorithm, we identified novel diagnostic markers for soft tissue sarcoma that distinguished between synovial sarcoma and MPNST, and that we validated using qRT-PCR in an independent series. Next, we identified prognostic genes that are strong predictors of disease outcome when used in a k-nearest neighbor algorithm. The prognostic genes were further validated in expression data from the French Sarcoma Group. One of these, HMMR, was validated in an independent series of leiomyosarcomas using immunohistochemistry on tissue micro array as a prognostic gene for disease-free interval. Furthermore, reconstruction of regulatory networks combined with data from the Connectivity Map showed, amongst others, that HDAC inhibitors could be a potential effective therapy for multiple soft tissue sarcoma subtypes. A viability assay with two HDAC inhibitors confirmed that both leiomyosarcoma and synovial sarcoma are sensitive to HDAC inhibition. In this study we identified novel diagnostic markers, prognostic markers and therapeutic leads from multiple soft tissue sarcoma gene expression datasets. Thus, machine learning algorithms are powerful new tools to improve our understanding of rare tumor entities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. 2013.
-
- Van Der Maaten L, Hinton G. Visualizing Data using t-SNE [Internet]. Journal of Machine Learning Research. 2008. http://www.jmlr.org/papers/volume9/vandermaaten08a/vandermaaten08a.pdf
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

