Crystal Structure and Conformational Dynamics of Pyrococcus furiosus Prolyl Oligopeptidase
- PMID: 30786206
- PMCID: PMC6714975
- DOI: 10.1021/acs.biochem.9b00031
Crystal Structure and Conformational Dynamics of Pyrococcus furiosus Prolyl Oligopeptidase
Abstract
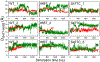
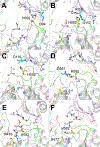
Enzymes in the prolyl oligopeptidase family possess unique structures and substrate specificities that are important for their biological activity and for potential biocatalytic applications. The crystal structures of Pyrococcus furiosus ( Pfu) prolyl oligopeptidase (POP) and the corresponding S477C mutant were determined to 1.9 and 2.2 Å resolution, respectively. The wild type enzyme crystallized in an open conformation, indicating that this state is readily accessible, and it contained bound chloride ions and a prolylproline ligand. These structures were used as starting points for molecular dynamics simulations of Pfu POP conformational dynamics. The simulations showed that large-scale domain opening and closing occurred spontaneously, providing facile substrate access to the active site. Movement of the loop containing the catalytically essential histidine into a conformation similar to those found in structures with fully formed catalytic triads also occurred. This movement was modulated by chloride binding, providing a rationale for experimentally observed activation of POP peptidase catalysis by chloride. Thus, the structures and simulations reported in this study, combined with existing biochemical data, provide a number of insights into POP catalysis.
Figures
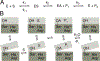





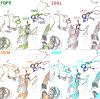

References
-
- Szeltner Z; Polgar L Structure, Function and Biological Relevance of Prolyl Oligopeptidase. Curr. Protein Pept. Sci. 2008, 9 (1), 96–107. - PubMed
-
- Rea D; Fülöp V Structure-Function Properties of Prolyl Oligopeptidase Family Enzymes. Cell Biochem. Biophys. 2006, 44 (3), 349–365. - PubMed
-
- Walter R; Shlank H; Glass JD; Schwartz IL; Kerenyi TD Leucylglycinamide Released From Oxytocin by Human Uterine Enzyme. Science 1971, 173 (3999), 827–829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

