High-Resolution Regulatory Maps Connect Vascular Risk Variants to Disease-Related Pathways
- PMID: 30786239
- PMCID: PMC8104016
- DOI: 10.1161/CIRCGEN.118.002353
High-Resolution Regulatory Maps Connect Vascular Risk Variants to Disease-Related Pathways
Erratum in
-
Correction to: High-Resolution Regulatory Maps Connect Vascular Risk Variants to Disease-Related Pathways.Circ Genom Precis Med. 2019 Aug;12(8):e000060. doi: 10.1161/HCG.0000000000000060. Epub 2019 Aug 20. Circ Genom Precis Med. 2019. PMID: 31430212 No abstract available.
-
Correction to: High-Resolution Regulatory Maps Connect Vascular Risk Variants to Disease-Related Pathways.Circ Genom Precis Med. 2021 Apr;14(2):e000081. doi: 10.1161/HCG.0000000000000081. Epub 2021 Mar 2. Circ Genom Precis Med. 2021. PMID: 33653079 Free PMC article. No abstract available.
-
Correction to: High-Resolution Regulatory Maps Connect Vascular Risk Variants to Disease-Related Pathways.Circ Genom Precis Med. 2021 Oct;14(5):e000087. doi: 10.1161/HCG.0000000000000087. Epub 2021 Oct 19. Circ Genom Precis Med. 2021. PMID: 34665644 No abstract available.
Abstract
Background: Genetic variant landscape of coronary artery disease is dominated by noncoding variants among which many occur within putative enhancers regulating the expression levels of relevant genes. It is crucial to assign the genetic variants to their correct genes both to gain insights into perturbed functions and better assess the risk of disease.
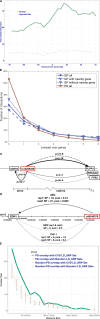
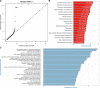
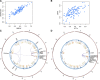
Methods: In this study, we generated high-resolution genomic interaction maps (≈750 bases) in aortic endothelial, smooth muscle cells and THP-1 (human leukemia monocytic cell line) macrophages stimulated with lipopolysaccharide using Hi-C coupled with sequence capture targeting 25 429 features, including variants associated with coronary artery disease. We also sequenced their transcriptomes and mapped putative enhancers using chromatin immunoprecipitation with an antibody against H3K27Ac.
Results: The regions interacting with promoters showed strong enrichment for enhancer elements and validated several previously known interactions and enhancers. We detected interactions for 727 risk variants obtained by genome-wide association studies and identified novel, as well as established genes and functions associated with cardiovascular diseases. We were able to assign potential target genes for additional 398 genome-wide association studies variants using haplotype information, thereby identifying additional relevant genes and functions. Importantly, we discovered that a subset of risk variants interact with multiple promoters and their expression levels were strongly correlated.
Conclusions: In summary, we present a catalog of candidate genes regulated by coronary artery disease-related variants and think that it will be an invaluable resource to further the investigation of cardiovascular pathologies and disease.
Keywords: coronary artery disease; gene; genomics; haplotype; inflammation.
Figures

References
-
- Reiner Z, et al. European Association for Cardiovascular Prevention & Rehabilitation. ESC/EAS Guidelines for the management of dyslipidaemias: the task force for the management of dyslipidaemias of the European Society of Cardiology (ESC) and the European Atherosclerosis Society (EAS). Eur Heart J. 2011;32:1769–1818. doi: 10.1093/eurheartj/ehr158. - PubMed
-
- Perk J, et al. European Association for Cardiovascular Prevention & Rehabilitation (EACPR) European guidelines on cardiovascular disease prevention in clinical practice (version 2012): the fifth joint task force of the European society of cardiology and other societies on cardiovascular disease prevention in clinical practice (constituted by representatives of nine societies and by invited experts). Int J Behav Med. 2012;19:403–488. doi: 10.1007/s12529-012-9242-5. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

