Determinants of Orexin Receptor Binding and Activation-A Molecular Dynamics Study
- PMID: 30786708
- PMCID: PMC6727383
- DOI: 10.1021/acs.jpcb.8b10220
Determinants of Orexin Receptor Binding and Activation-A Molecular Dynamics Study
Abstract
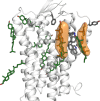
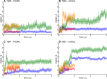
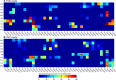
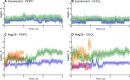
We assess the stability of two previously suggested binding modes for the neuropeptide orexin-A in the OX2 receptor through extensive molecular dynamics simulations. As the activation determinants of the receptor remain unknown, we simulated an unliganded receptor and two small-molecular ligands, the antagonist suvorexant and the agonist Nag26 for comparison. Each system was simulated in pure POPC membrane as well as in the 25% cholesterol-POPC membrane. In total, we carried out 36 μs of simulations. Through this set of simulations, we report a stable binding mode for the C-terminus of orexin-A. In addition, we suggest interactions that would promote orexin receptor activation, as well as others that would stabilize the inactive state.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- Cox C. D.; Breslin M. J.; Whitman D. B.; Schreier J. D.; McGaughey G. B.; Bogusky M. J.; Roecker A. J.; Mercer S. P.; Bednar R. a; Lemaire W.; et al. Discovery of the Dual Orexin Receptor Antagonist [(7R)-4-(5-Chloro-1,3-Benzoxazol-2-Yl)-7-Methyl-1,4-Diazepan-1-Yl][5-Methyl-2-(2H-1,2,3-Triazol-2-Yl)Phenyl]Methanone (MK-4305) for the Treatment of Insomnia. J. Med. Chem. 2010, 53, 5320–5332. 10.1021/jm100541c. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

