Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre
- PMID: 30786920
- PMCID: PMC6381711
- DOI: 10.1186/s40478-019-0668-8
Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre
Abstract
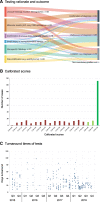
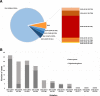
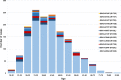
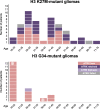
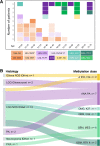
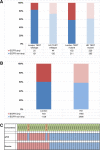
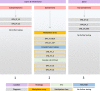
The introduction of the classification of brain tumours based on their DNA methylation profile has significantly changed the diagnostic approach for cases with ambiguous histology, non-informative or contradictory molecular profiles or for entities where methylation profiling provides useful information for patient risk stratification, for example in medulloblastoma and ependymoma. We present our experience that combines a conventional molecular diagnostic approach with the complementary use of a DNA methylation-based classification tool, for adult brain tumours originating from local as well as national referrals. We report the frequency of IDH mutations in a large cohort of nearly 1550 patients, EGFR amplifications in almost 1900 IDH-wildtype glioblastomas, and histone mutations in 70 adult gliomas. We demonstrate how additional methylation-based classification has changed and improved our diagnostic approach. Of the 325 cases referred for methylome testing, 179 (56%) had a calibrated score of 0.84 and higher and were included in the evaluation. In these 179 samples, the diagnosis was changed in 45 (25%), refined in 86 (48%) and confirmed in 44 cases (25%). In addition, the methylation arrays contain copy number information that usefully complements the methylation profile. For example, EGFR amplification which is 95% concordant with our Real-Time PCR-based copy number assays. We propose here a diagnostic algorithm that integrates histology, conventional molecular tests and methylation arrays.
Keywords: Adult brain tumours; BRAF; Brain tumour classification; DKFZ classifier; Ependymoma; Glioma; H3 K27M; Histone mutation; IDH1; IDH2; Illumina array; Methylation array; Methylation classifier; Molecular diagnostics.
Conflict of interest statement
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Holsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Bruck W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hanggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Muhleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Muller HL, Rutkowski S, von Hoff K, Fruhwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu CM, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blumcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schuller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A, Pfister SM. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. - DOI - PMC - PubMed
-
- Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G, Pusch S, Mechtersheimer G, Zentgraf H, von Deimling A. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. - DOI - PubMed
-
- Capper D, Stichel D, Sahm F, Jones DTW, Schrimpf D, Sill M, Schmid S, Hovestadt V, Reuss DE, Koelsche C, Reinhardt A, Wefers AK, Huang K, Sievers P, Ebrahimi A, Scholer A, Teichmann D, Koch A, Hanggi D, Unterberg A, Platten M, Wick W, Witt O, Milde T, Korshunov A, Pfister SM, von Deimling A. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136:181–210. doi: 10.1007/s00401-018-1879-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

