Integrated systems approach defines the antiviral pathways conferring protection by the RV144 HIV vaccine
- PMID: 30787294
- PMCID: PMC6382801
- DOI: 10.1038/s41467-019-08854-2
Integrated systems approach defines the antiviral pathways conferring protection by the RV144 HIV vaccine
Abstract
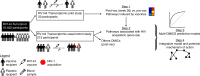
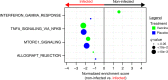
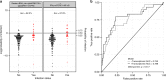
The RV144 vaccine trial showed reduced risk of HIV-1 acquisition by 31.2%, although mechanisms that led to protection remain poorly understood. Here we identify transcriptional correlates for reduced HIV-1 acquisition after vaccination. We assess the transcriptomic profile of blood collected from 223 participants and 40 placebo recipients. Pathway-level analysis of HIV-1 negative vaccinees reveals that type I interferons that activate the IRF7 antiviral program and type II interferon-stimulated genes implicated in antigen-presentation are both associated with a reduced risk of HIV-1 acquisition. In contrast, genes upstream and downstream of NF-κB, mTORC1 and host genes required for viral infection are associated with an increased risk of HIV-1 acquisition among vaccinees and placebo recipients, defining a vaccine independent association with HIV-1 acquisition. Our transcriptomic analysis of RV144 trial samples identifies IRF7 as a mediator of protection and the activation of mTORC1 as a correlate of the risk of HIV-1 acquisition.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

