Targeted editing of the PSIP1 gene encoding LEDGF/p75 protects cells against HIV infection
- PMID: 30787394
- PMCID: PMC6382798
- DOI: 10.1038/s41598-019-38718-0
Targeted editing of the PSIP1 gene encoding LEDGF/p75 protects cells against HIV infection
Abstract
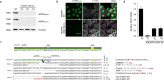
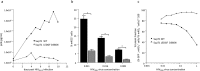
To fulfill a productive infection cycle the human immunodeficiency virus (HIV) relies on host-cell factors. Interference with these co-factors holds great promise in protecting cells against HIV infection. LEDGF/p75, encoded by the PSIP1 gene, is used by the integrase (IN) protein in the pre-integration complex of HIV to bind host-cell chromatin facilitating proviral integration. LEDGF/p75 depletion results in defective HIV replication. However, as part of its cellular function LEDGF/p75 tethers cellular proteins to the host-cell genome. We used site-specific editing of the PSIP1 locus using CRISPR/Cas to target the aspartic acid residue in position 366 and mutated it to asparagine (D366N) to disrupt the interaction with HIV IN but retain LEDGF/p75 cellular function. The resulting cell lines demonstrated successful disruption of the LEDGF/p75 HIV-IN interface without affecting interaction with cellular binding partners. In line with LEDGF/p75 depleted cells, D366N cells did not support HIV replication, in part due to decreased integration efficiency. In addition, we confirm the remaining integrated provirus is more silent. Taken together, these results support the potential of site-directed CRISPR/Cas9 mediated knock-in to render cells more resistant to HIV infection and provides an additional strategy to protect patient-derived T-cells against HIV-1 infection as part of cell-based therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

