De novo origins of multicellularity in response to predation
- PMID: 30787483
- PMCID: PMC6382799
- DOI: 10.1038/s41598-019-39558-8
De novo origins of multicellularity in response to predation
Abstract
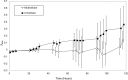
The transition from unicellular to multicellular life was one of a few major events in the history of life that created new opportunities for more complex biological systems to evolve. Predation is hypothesized as one selective pressure that may have driven the evolution of multicellularity. Here we show that de novo origins of simple multicellularity can evolve in response to predation. We subjected outcrossed populations of the unicellular green alga Chlamydomonas reinhardtii to selection by the filter-feeding predator Paramecium tetraurelia. Two of five experimental populations evolved multicellular structures not observed in unselected control populations within ~750 asexual generations. Considerable variation exists in the evolved multicellular life cycles, with both cell number and propagule size varying among isolates. Survival assays show that evolved multicellular traits provide effective protection against predation. These results support the hypothesis that selection imposed by predators may have played a role in some origins of multicellularity.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Buss, L. W. The Evolution of Individuality. (Princeton University Press, 1987).
-
- Grosberg RK, Strathmann RR. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 2007;38:621–654. doi: 10.1146/annurev.ecolsys.36.102403.114735. - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

