The evolution of phenotypic plasticity when environments fluctuate in time and space
- PMID: 30788139
- PMCID: PMC6369965
- DOI: 10.1002/evl3.100
The evolution of phenotypic plasticity when environments fluctuate in time and space
Abstract
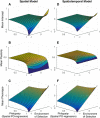
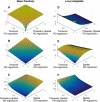
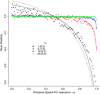
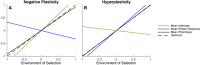
Most theoretical studies have explored the evolution of plasticity when the environment, and therefore the optimal trait value, varies in time or space. When the environment varies in time and space, we show that genetic adaptation to Markovian temporal fluctuations depends on the between-generation autocorrelation in the environment in exactly the same way that genetic adaptation to spatial fluctuations depends on the probability of philopatry. This is because both measure the correlation in parent-offspring environments and therefore the effectiveness of a genetic response to selection. If the capacity to genetically respond to selection is stronger in one dimension (e.g., space), then plasticity mainly evolves in response to fluctuations in the other dimension (e.g., time). If the relationships between the environments of development and selection are the same in time and space, the evolved plastic response to temporal fluctuations is useful in a spatial context and genetic differentiation in space is reduced. However, if the relationships between the environments of development and selection are different, the optimal level of plasticity is different in the two dimensions. In this case, the plastic response that evolves to cope with temporal fluctuations may actually be maladaptive in space, resulting in the evolution of hyperplasticity or negative plasticity. These effects can be mitigated by spatial genetic differentiation that acts in opposition to plasticity resulting in counter-gradient variation. These results highlight the difficulty of making space-for-time substitutions in empirical work but identify the key parameters that need to be measured in order to test whether space-for-time substitutions are likely to be valid.
Keywords: Counter‐gradient variation; environmental heterogeneity; hyperplasticity; local adaptation; phenotypic plasticity; space; time.
Figures
References
-
- Blanquart, F. , Gandon S., and Nuismer S. L.. 2012. The effects of migration and drift on local adaptation to a heterogeneous environment. J. Evol. Biol. 25: 1351–1363. - PubMed
-
- Bull, J . 1987. Evolution of phenotypic variance. Evolution 41: 303–315. - PubMed
-
- van Buskirk, J . 2017. Spatially heterogeneous selection in nature favors phenotypic plasticity in anuran larvae. Evolution 71: 1670–1685. - PubMed
-
- Cenzer, M. L . 2017. Maladaptive plasticity masks the effects of natural selection in the red‐shouldered soapberry bug. Am. Nat. 190: 521–533. - PubMed
-
- Chevin, L.‐M. , and Lande R.. 2015. Evolution of environmental cues for phenotypic plasticity. Evolution 69: 2767–2775. - PubMed

